8XFQ
 
 | |
8XFR
 
 | |
4I86
 
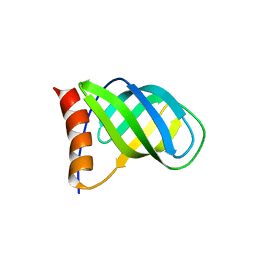 | | Crystal structure of PilZ domain of CeSA from cellulose synthesizing bacterium | | 分子名称: | Cellulose synthase 1 | | 著者 | Fujiwara, T, Komoda, K, Sakurai, N, Tanaka, I, Yao, M. | | 登録日 | 2012-12-03 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.098 Å) | | 主引用文献 | The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A
Biochem.Biophys.Res.Commun., 431, 2013
|
|
8JA4
 
 | | Structure of the alginate epimerase/lyase | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | 著者 | Fujiwara, T. | | 登録日 | 2023-05-05 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural basis for the minimal bifunctional alginate epimerase AlgE3 from Azotobacter chroococcum.
Febs Lett., 2024
|
|
8JA6
 
 | |
8JAZ
 
 | |
7F81
 
 | | Structure of the bacterial cellulose synthase subunit Z from Enterobacter sp. CJF-002 | | 分子名称: | GLYCEROL, Glucanase, S,R MESO-TARTARIC ACID | | 著者 | Fujiwara, T, Fujishima, A, Yao, M. | | 登録日 | 2021-06-30 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7F82
 
 | | Structure of the bacterial cellulose synthase subunit Z in complex with cellooligosaccharides from Enterobacter sp. CJF-002 | | 分子名称: | Glucanase, S,R MESO-TARTARIC ACID, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | 著者 | Fujiwara, T, Fujishima, A, Yao, M. | | 登録日 | 2021-06-30 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Acta Crystallogr.,Sect.D, 78, 2022
|
|
3VW5
 
 | |
3WKH
 
 | | Crystal structure of cellobiose 2-epimerase in complex with epilactose | | 分子名称: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | 著者 | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | 登録日 | 2013-10-21 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.644 Å) | | 主引用文献 | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WKG
 
 | | Crystal structure of cellobiose 2-epimerase in complex with glucosylmannose | | 分子名称: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | 著者 | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | 登録日 | 2013-10-21 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WKI
 
 | | Crystal structure of cellobiose 2-epimerase in complex with cellobiitol | | 分子名称: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | 著者 | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | 登録日 | 2013-10-21 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.191 Å) | | 主引用文献 | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WKF
 
 | | Crystal structure of cellobiose 2-epimerase | | 分子名称: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION | | 著者 | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | 登録日 | 2013-10-21 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.743 Å) | | 主引用文献 | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
6M0Q
 
 | | Hydroxylamine oxidoreductase from Nitrosomonas europaea | | 分子名称: | Aerobic hydroxylamine oxidoreductase, DI(HYDROXYETHYL)ETHER, HEME C, ... | | 著者 | Fujiwara, T, Fujimoto, Z, Nishigaya, Y, Yamazaki, T. | | 登録日 | 2020-02-22 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Juglone, a plant-derived 1,4-naphthoquinone, binds to hydroxylamine oxidoreductase and inhibits the electron transfer to cytochrome c 554.
Appl.Environ.Microbiol., 89, 2023
|
|
6M0P
 
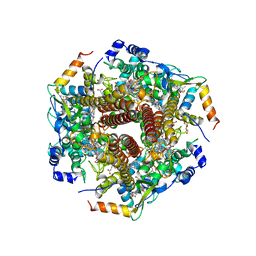 | | Hydroxylamine oxidoreductase in complex with juglone | | 分子名称: | 5-hydroxynaphthalene-1,4-dione, Aerobic hydroxylamine oxidoreductase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Fujiwara, T, Fujimoto, Z, Nishigaya, Y, Yamazaki, T. | | 登録日 | 2020-02-22 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Juglone, a plant-derived 1,4-naphthoquinone, binds to hydroxylamine oxidoreductase and inhibits the electron transfer to cytochrome c 554.
Appl.Environ.Microbiol., 89, 2023
|
|
2E8D
 
 | |
8HEW
 
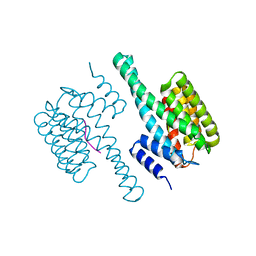 | | Potato 14-3-3 St14f | | 分子名称: | 14-3-3 protein, StFDL1 peptide | | 著者 | Taoka, K, Kawahara, I, Shinya, S, Harada, K, Muranaka, T, Furuita, K, Nakagawa, A, Fujiwara, T, Tsuji, H, Kojima, C. | | 登録日 | 2022-11-08 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Multifunctional chemical inhibitors of the florigen activation complex discovered by structure-based high-throughput screening.
Plant J., 112, 2022
|
|
4U5X
 
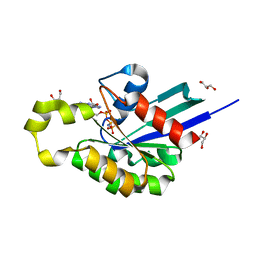 | | Structure of plant small GTPase OsRac1 complexed with the non-hydrolyzable GTP analog GMPPNP | | 分子名称: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Ohki, I, Kosami, K, Fujiwara, T, Nakagawa, A, Shimamoto, K, Kojima, C. | | 登録日 | 2014-07-25 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Crystal Structure of the Plant Small GTPase OsRac1 Reveals Its Mode of Binding to NADPH Oxidase
J.Biol.Chem., 289, 2014
|
|
6LLQ
 
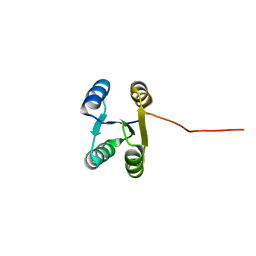 | | Solution NMR structure of de novo Rossmann2x2 fold with most of the core mutated to valine, R2x2_VAL88 | | 分子名称: | VAL88 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Koga, R, Yamamoto, M, Kosugi, T, Koga, N. | | 登録日 | 2019-12-23 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Robust folding of a de novo designed ideal protein even with most of the core mutated to valine.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1R77
 
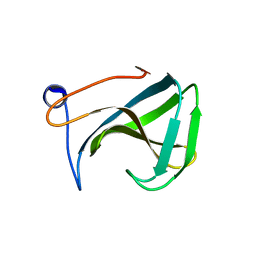 | | Crystal structure of the cell wall targeting domain of peptidylglycan hydrolase ALE-1 | | 分子名称: | Cell Wall Targeting Domain of Glycylglycine Endopeptidase ALE-1 | | 著者 | Lu, J.Z, Fujiwara, T, Komatsuzawa, H, Sugai, M, Sakon, J. | | 登録日 | 2003-10-20 | | 公開日 | 2005-04-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Cell Wall-targeting Domain of Glycylglycine Endopeptidase Distinguishes among Peptidoglycan Cross-bridges.
J.Biol.Chem., 281, 2006
|
|
1QS4
 
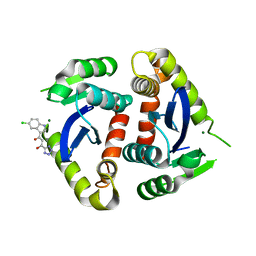 | | Core domain of HIV-1 integrase complexed with Mg++ and 1-(5-chloroindol-3-yl)-3-hydroxy-3-(2H-tetrazol-5-yl)-propenone | | 分子名称: | 1-(5-CHLOROINDOL-3-YL)-3-HYDROXY-3-(2H-TETRAZOL-5-YL)-PROPENONE, MAGNESIUM ION, PROTEIN (HIV-1 INTEGRASE (E.C.2.7.7.49)) | | 著者 | Goldgur, Y, Craigie, R, Fujiwara, T, Yoshinaga, T, Davies, D.R. | | 登録日 | 1999-06-25 | | 公開日 | 1999-11-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5WS3
 
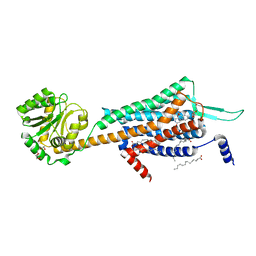 | | Crystal structures of human orexin 2 receptor bound to the selective antagonist EMPA determined by serial femtosecond crystallography at SACLA | | 分子名称: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | 著者 | Suno, R, Kimura, K, Nakane, T, Yamashita, K, Wang, J, Fujiwara, T, Yamanaka, Y, Im, D, Tsujimoto, H, Sasanuma, M, Horita, S, Hirokawa, T, Nango, E, Tono, K, Kameshima, T, Hatsui, T, Joti, Y, Yabashi, M, Shimamoto, K, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | 登録日 | 2016-12-05 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA.
Structure, 26, 2018
|
|
5JR0
 
 | |
2ND5
 
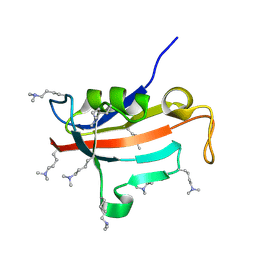 | | Lysine dimethylated FKBP12 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP1A | | 著者 | Hattori, Y, Sebera, J, Sychrovsky, V, Furuita, K, Sugiki, T, Ohki, I, Ikegami, T, Kobayashi, N, Tanaka, Y, Fujiwara, T, Kojima, C. | | 登録日 | 2016-05-05 | | 公開日 | 2017-05-17 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Observation of Protein Surface Salt Bridges at Neutral pH
To be Published
|
|
7BQN
 
 | | Solution NMR structure of fold-C Rei; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Rei | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
