5M1S
 
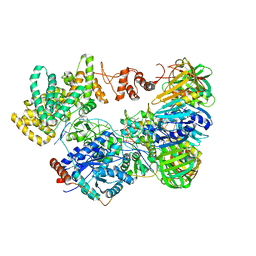 | | Cryo-EM structure of the E. coli replicative DNA polymerase-clamp-exonuclase-theta complex bound to DNA in the editing mode | | 分子名称: | DNA Primer Strand, DNA Template Strand, DNA polymerase III subunit alpha, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2016-10-10 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Self-correcting mismatches during high-fidelity DNA replication.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5FKU
 
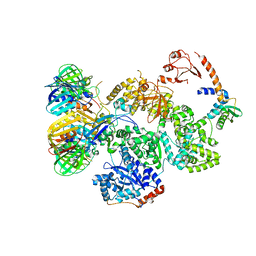 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | 分子名称: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2015-10-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8.34 Å) | | 主引用文献 | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
3LRK
 
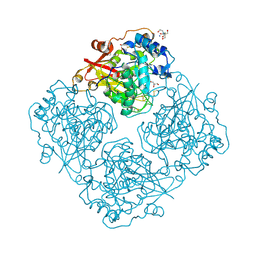 | | Structure of alfa-galactosidase (MEL1) from Saccharomyces cerevisiae | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | 登録日 | 2010-02-11 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
3LRM
 
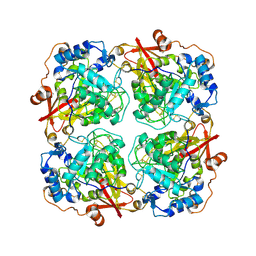 | | Structure of alfa-galactosidase from Saccharomyces cerevisiae with raffinose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | 登録日 | 2010-02-11 | | 公開日 | 2010-06-30 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
3LRL
 
 | | Structure of alfa-galactosidase (MEL1) from Saccharomyces cerevisiae with melibiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | 登録日 | 2010-02-11 | | 公開日 | 2010-06-30 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
5FKV
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | 分子名称: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2015-10-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8.04 Å) | | 主引用文献 | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FKW
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon) | | 分子名称: | DNA POLYMERASE III ALPHA, DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2015-10-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
7AI7
 
 | | MutS in Intermediate state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*CP*TP*TP*AP*GP*CP*TP*TP*AP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*TP*AP*AP*CP*TP*AP*AP*G)-3'), ... | | 著者 | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | 登録日 | 2020-09-26 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AIC
 
 | | MutS-MutL in clamp state (kinked clamp domain) | | 分子名称: | DNA (30-MER), DNA mismatch repair protein MutL, DNA mismatch repair protein MutS, ... | | 著者 | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | 登録日 | 2020-09-26 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AI6
 
 | | MutS in mismatch bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (25-MER), DNA mismatch repair protein MutS | | 著者 | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | 登録日 | 2020-09-26 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AIB
 
 | | MutS-MutL in clamp state | | 分子名称: | DNA (30-MER), DNA mismatch repair protein MutL, DNA mismatch repair protein MutS, ... | | 著者 | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | 登録日 | 2020-09-26 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AI5
 
 | | MutS in Scanning state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*AP*AP*TP*TP*CP*GP*CP*CP*CP*TP*AP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*TP*AP*GP*GP*GP*CP*GP*AP*AP*TP*TP*GP*GP*GP*TP*AP*CP*CP*G)-3'), ... | | 著者 | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | 登録日 | 2020-09-26 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3OB8
 
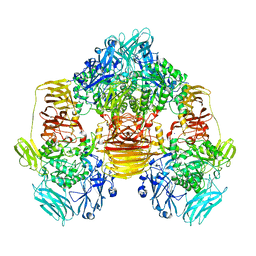 | | Structure of the beta-galactosidase from Kluyveromyces lactis in complex with galactose | | 分子名称: | Beta-galactosidase, MAGNESIUM ION, MANGANESE (II) ION, ... | | 著者 | Fernandez-Leiro, R, Pereira-Rodriguez, A, Becerra, M, Gonzalez-Siso, I, Cerdan, M.E, Sanz-Aparicio, J. | | 登録日 | 2010-08-06 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of specificity in tetrameric Kluyveromyces lactis beta-galactosidase.
J.Struct.Biol., 177, 2012
|
|
3OBA
 
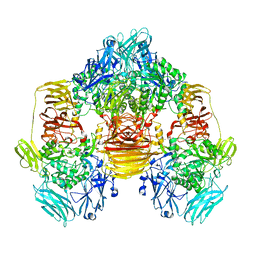 | | Structure of the beta-galactosidase from Kluyveromyces lactis | | 分子名称: | Beta-galactosidase, GLYCEROL, MANGANESE (III) ION | | 著者 | Fernandez-Leiro, R, Pereira-Rodriguez, A, Becerra, M, Gonzalez-Siso, I, Cerdan, M.E, Sanz-Aparicio, J. | | 登録日 | 2010-08-06 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural basis of specificity in tetrameric Kluyveromyces lactis beta-galactosidase.
J.Struct.Biol., 177, 2012
|
|
6S6Z
 
 | | Structure of beta-Galactosidase from Thermotoga maritima | | 分子名称: | Beta-galactosidase, MAGNESIUM ION | | 著者 | Miguez-Amil, S, Jimenez-Ortega, E, Ramirez Escudero, M, Sanz-Aparicio, J, Fernandez-Leiro, R. | | 登録日 | 2019-07-04 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2 Å) | | 主引用文献 | The cryo-EM Structure ofThermotoga maritimabeta-Galactosidase: Quaternary Structure Guides Protein Engineering.
Acs Chem.Biol., 15, 2020
|
|
5IDE
 
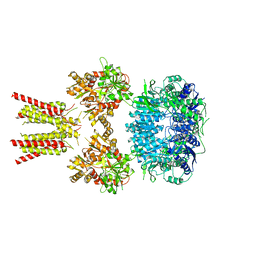 | | Cryo-EM structure of GluA2/3 AMPA receptor heterotetramer (model I) | | 分子名称: | Glutamate receptor 2, Glutamate receptor 3 | | 著者 | Herguedas, B, Garcia-Nafria, J, Fernandez-Leiro, R, Greger, I.H. | | 登録日 | 2016-02-24 | | 公開日 | 2016-03-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (8.25 Å) | | 主引用文献 | Structure and organization of heteromeric AMPA-type glutamate receptors.
Science, 352, 2016
|
|
5IDF
 
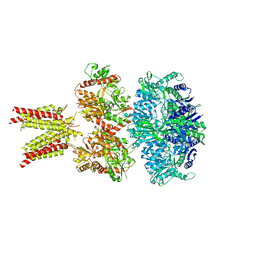 | | Cryo-EM structure of GluA2/3 AMPA receptor heterotetramer (model II) | | 分子名称: | Glutamate receptor 2, Glutamate receptor 3 | | 著者 | Herguedas, B, Garcia-Nafria, J, Fernandez-Leiro, R, Greger, I.H. | | 登録日 | 2016-02-24 | | 公開日 | 2016-03-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (10.31 Å) | | 主引用文献 | Structure and organization of heteromeric AMPA-type glutamate receptors.
Science, 352, 2016
|
|
7AS4
 
 | | Recombinant human gTuRC | | 分子名称: | Actin, cytoplasmic 1, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Serna, M, Fernandez-Leiro, R, Llorca, O. | | 登録日 | 2020-10-26 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.13 Å) | | 主引用文献 | Assembly of the asymmetric human gamma-tubulin ring complex by RUVBL1-RUVBL2 AAA ATPase.
Sci Adv, 6, 2020
|
|
8AG3
 
 | |
8AG4
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | 分子名称: | Protein C10, X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | 著者 | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | 登録日 | 2022-07-19 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.46 Å) | | 主引用文献 | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
8AG5
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | 分子名称: | Ku70-Xrcc6, Protein C10, X-ray repair cross-complementing protein 5 | | 著者 | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | 登録日 | 2022-07-19 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
6QI9
 
 | | Truncated human R2TP complex, structure 4 (ADP-empty) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | 著者 | Munoz-Hernandez, H, Rodriguez, C.F, Llorca, O. | | 登録日 | 2019-01-18 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.63 Å) | | 主引用文献 | Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6QI8
 
 | | Truncated human R2TP complex, structure 3 (ADP-filled) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | 著者 | Munoz-Hernandez, H, Rodriguez, C.F, Llorca, O. | | 登録日 | 2019-01-18 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6HPO
 
 | |
6HYC
 
 | |
