5J7M
 
 | | Crystal structure of Cupin 2 conserved barrel domain protein from Kribbella flavida DSM 17836 | | 分子名称: | ACETATE ION, Cupin 2 conserved barrel domain protein, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Chang, C, Cuff, M, Chhor, G, Endres, M, Joachimiak, A. | | 登録日 | 2016-04-06 | | 公開日 | 2016-04-27 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystal structure of Cupin 2 conserved barrel domain protein from Kribbella flavida DSM 17836
To Be Published
|
|
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | 分子名称: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2023-02-14 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
8UW6
 
 | | Acetylornithine deacetylase from Escherichia coli, di-zinc form. | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetylornithine deacetylase, ... | | 著者 | Osipiuk, J, Endres, M, Kelley, E, Becker, D.P, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2023-11-06 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Acetylornithine deacetylase from Escherichia coli, di-zinc form.
To Be Published
|
|
9BZB
 
 | | Crystal structure of metallo-hydrolase-like_MBL-fold protein from Salmonella typhimurium LT2 | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, SULFATE ION, ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2024-05-24 | | 公開日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of metallo-hydrolase-like_MBL-fold protein from Salmonella typhimurium LT2
To Be Published
|
|
4NPX
 
 | | Structure of hypothetical protein Cj0539 from Campylobacter jejuni | | 分子名称: | Putative uncharacterized protein | | 著者 | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Adkins, J.N, Endres, M, Nissen, M, Konkel, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | 登録日 | 2013-11-22 | | 公開日 | 2014-01-01 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structure of hypothetical protein Cj0539 from Campylobacter jejuni
To be Published
|
|
6DT3
 
 | | 1.2 Angstrom Resolution Crystal Structure of Nucleoside Triphosphatase NudI from Klebsiella pneumoniae in Complex with HEPES | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nucleoside triphosphatase NudI | | 著者 | Minasov, G, Shuvalova, L, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-06-15 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6DVV
 
 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD and Mn2+. | | 分子名称: | 6-phospho-alpha-glucosidase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-06-25 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6DXN
 
 | | 1.95 Angstrom Resolution Crystal Structure of DsbA Disulfide Interchange Protein from Klebsiella pneumoniae. | | 分子名称: | TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein | | 著者 | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-06-29 | | 公開日 | 2018-07-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6X9Y
 
 | | The crystal structure of a Beta-lactamase from Escherichia coli CFT073 | | 分子名称: | Beta-lactamase, GLYCEROL, S,R MESO-TARTARIC ACID, ... | | 著者 | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure of a Beta-lactamase from Escherichia coli CFT073
To Be Published
|
|
6XG1
 
 | | Class C beta-lactamase from Escherichia coli | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase | | 著者 | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-16 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Class C beta-lactamase from Escherichia coli
To Be Published
|
|
6XG3
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, at room temperature | | 分子名称: | CHLORIDE ION, Non-structural protein 3, PHOSPHATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-16 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
6XFS
 
 | | Class C beta-lactamase from Escherichia coli in complex with Tazobactam | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-16 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Class C beta-lactamase from Escherichia coli in complex with Tazobactam
To Be Published
|
|
6XPM
 
 | | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus with microfluidics crystals at room temperature | | 分子名称: | Lysophospholipase L1, SODIUM ION | | 著者 | Kim, Y, Johnson, J, Welk, L, Endres, M, Levens, A, Sherrell, D.A, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus with microfluidics crystals at room temperature
To Be Published
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | 分子名称: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | 著者 | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-10-03 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5IXP
 
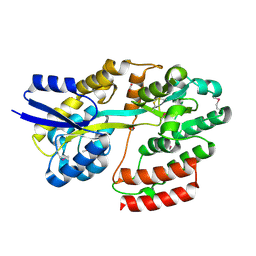 | | Crystal structure of Extracellular solute-binding protein family 1 | | 分子名称: | Extracellular solute-binding protein family 1, FORMIC ACID | | 著者 | Chang, C, Cuff, M, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-03-23 | | 公開日 | 2016-03-30 | | 最終更新日 | 2016-08-17 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystal structure of Extracellular solute-binding protein family 1
To Be Published
|
|
4X2R
 
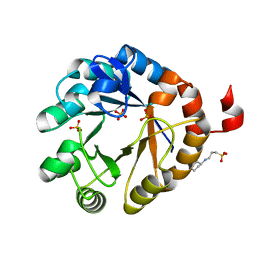 | | Crystal structure of PriA from Actinomyces urogenitalis | | 分子名称: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, PHOSPHATE ION | | 著者 | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-26 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Evolution of substrate specificity in a retained enzyme driven by gene loss.
Elife, 6, 2017
|
|
4X9S
 
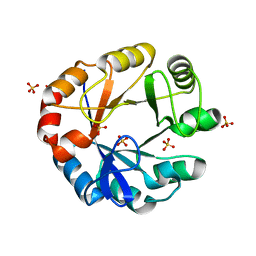 | | CRYSTAL STRUCTURE OF HISAP FROM STREPTOMYCES SP. MG1 | | 分子名称: | Phosphoribosyl isomerase A, SULFATE ION | | 著者 | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-12-11 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4XR9
 
 | | Crystal structure of CalS8 from Micromonospora echinospora cocrystallized with NAD and TDP-glucose | | 分子名称: | CalS8, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-01-20 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of CalS8 from Micromonospora echinospora
To Be Published
|
|
4XED
 
 | | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20 | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Peptidase M14, ... | | 著者 | Michalska, K, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-12-23 | | 公開日 | 2015-05-13 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20
To Be Published
|
|
4XRR
 
 | | Crystal structure of cals8 from micromonospora echinospora (P294S mutant) | | 分子名称: | CalS8, GLYCEROL | | 著者 | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-01-21 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural Characterization of CalS8, a TDP-alpha-D-Glucose Dehydrogenase Involved in Calicheamicin Aminodideoxypentose Biosynthesis.
J. Biol. Chem., 290, 2015
|
|
4XVO
 
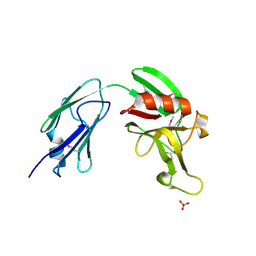 | | L,D-transpeptidase from Mycobacterium smegmatis | | 分子名称: | L,D-transpeptidase, PHOSPHATE ION | | 著者 | Osipiuk, J, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-01-27 | | 公開日 | 2015-02-11 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | L,D-transpeptidase from Mycobacterium smegmatis
to be published
|
|
4MNR
 
 | | Crystal Structure of D,D-Transpeptidase Domain of Peptidoglycan Glycosyltransferase from Eggerthella lenta | | 分子名称: | ACETIC ACID, MAGNESIUM ION, Peptidoglycan glycosyltransferase | | 著者 | Kim, Y, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-09-11 | | 公開日 | 2013-09-25 | | 実験手法 | X-RAY DIFFRACTION (1.653 Å) | | 主引用文献 | Crystal Structure of D,D-Transpeptidase Domain of Peptidoglycan Glycosyltransferase from Eggerthella lenta
To be Published
|
|
4N03
 
 | | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata | | 分子名称: | 1,2-ETHANEDIOL, ABC-type branched-chain amino acid transport systems periplasmic component-like protein, PALMITIC ACID | | 著者 | Osipiuk, J, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-09-30 | | 公開日 | 2013-10-16 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata
To be Published
|
|
4MVE
 
 | |
4N01
 
 | |
