2KI3
 
 | |
6XO3
 
 | |
6XN6
 
 | |
6XPA
 
 | |
6XOJ
 
 | |
6X1W
 
 | | Structure of pHis Fab (SC56-2) in complex with pHis mimetic peptide | | 分子名称: | ACLYana-3-pTza peptide, SC56-2 Heavy chain, SC56-2 Light chain | | 著者 | Kalagiri, R, Stanfield, R, Wilson, I.A, Hunter, T. | | 登録日 | 2020-05-19 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6X1U
 
 | | Structure of pHis Fab (SC39-4) in complex with pHis mimetic peptide | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, ACLYana-3-pTza peptide, ... | | 著者 | Kalagiri, R, Stanfield, R, Wilson, I.A, Hunter, T. | | 登録日 | 2020-05-19 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.641 Å) | | 主引用文献 | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6X1V
 
 | | Structure of pHis Fab (SC44-8) in complex with pHis mimetic peptide | | 分子名称: | 1,2-ETHANEDIOL, ACLYana-3-pTza peptide, SC44-8 Heavy chain, ... | | 著者 | Kalagiri, R, Stanfield, R, Wilson, I.A, Hunter, T. | | 登録日 | 2020-05-19 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6X1S
 
 | | Structure of pHis Fab (SC1-1) in complex with pHis mimetic peptide | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, NM23-1-pTza peptide, SC1-1 Heavy chain, ... | | 著者 | Kalagiri, R, Stanfield, R.L, Wilson, I.A, Hunter, T. | | 登録日 | 2020-05-19 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6X1T
 
 | | Structure of pHis Fab (SC50-3) in complex with pHis mimetic peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kalagiri, R, Stanfield, R.L, Wilson, I.A, Hunter, T. | | 登録日 | 2020-05-19 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3PA7
 
 | | Crystal structure of FKBP from plasmodium vivax in complex with tetrapeptide ALPF | | 分子名称: | 4-mer Peptide ALPF, 70 kDa peptidylprolyl isomerase, putative | | 著者 | Balakrishna, A.M, Alag, R, Yoon, H.S. | | 登録日 | 2010-10-18 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structural insights into substrate binding by PvFKBP35, a peptidylprolyl cis-trans isomerase from the human malarial parasite Plasmodium vivax
EUKARYOTIC CELL, 12, 2013
|
|
3IHZ
 
 | | Crystal structure of the FK506 binding domain of Plasmodium vivax FKBP35 in complex with FK506 | | 分子名称: | 70 kDa peptidylprolyl isomerase, putative, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | 著者 | Qureshi, I.A, Alag, R, Yoon, H.S, Lescar, J. | | 登録日 | 2009-07-31 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | NMR and crystallographic structures of the FK506 binding domain of human malarial parasite Plasmodium vivax FKBP35
Protein Sci., 19, 2010
|
|
2VN1
 
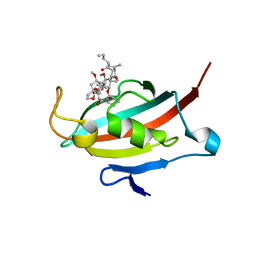 | | Crystal structure of the FK506-binding domain of Plasmodium falciparum FKBP35 in complex with FK506 | | 分子名称: | 70 KDA PEPTIDYLPROLYL ISOMERASE, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | 著者 | Kotaka, M, Alag, R, Ye, H, Preiser, P.R, Yoon, H.S, Lescar, J. | | 登録日 | 2008-01-30 | | 公開日 | 2008-05-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal Structure of the Fk506 Binding Domain of Plasmodium Falciparum Fkbp35 in Complex with Fk506.
Biochemistry, 47, 2008
|
|
4U9G
 
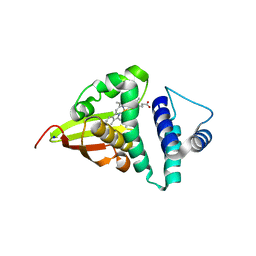 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)CO ligation state, Q154A/Q155A/K156A mutant | | 分子名称: | CARBON MONOXIDE, NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | 登録日 | 2014-08-06 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9K
 
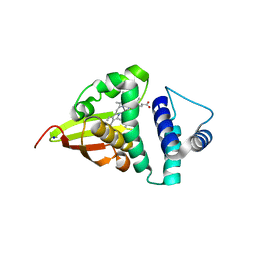 | | Crystal structure of an H-NOX protein from S. oneidensis in the Mn(II)NO ligation state, Q154A/Q155A/K156A mutant | | 分子名称: | MANGANESE PROTOPORPHYRIN IX, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | 著者 | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | 登録日 | 2014-08-06 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U99
 
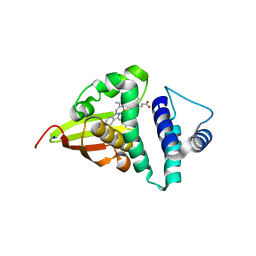 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II) ligation state, Q154A/Q155A/K156A mutant | | 分子名称: | NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | 著者 | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | 登録日 | 2014-08-05 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9J
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Mn(II) ligation state, Q154A/Q155A/K156A mutant | | 分子名称: | MANGANESE PROTOPORPHYRIN IX, NO-binding heme-dependent sensor protein, SODIUM ION, ... | | 著者 | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | 登録日 | 2014-08-06 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9B
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)NO ligation state | | 分子名称: | GLYCEROL, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | 著者 | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | 登録日 | 2014-08-05 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2FBN
 
 | | Plasmodium falciparum putative FK506-binding protein PFL2275c, C-terminal TPR-containing domain | | 分子名称: | 70 kDa peptidylprolyl isomerase, putative | | 著者 | Dong, A, Lew, J, Koeieradzki, I, Sundararajan, E, Melone, M, Wasney, G, Zhao, Y, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | 登録日 | 2005-12-09 | | 公開日 | 2006-01-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Crystallographic structure of the tetratricopeptide repeat domain of Plasmodium falciparum FKBP35 and its molecular interaction with Hsp90 C-terminal pentapeptide.
Protein Sci., 18, 2009
|
|
4ITZ
 
 | |
3NI6
 
 | |
8HGA
 
 | |
8HIA
 
 | |
5J6S
 
 | | Crystal structure of Endoplasmic Reticulum Aminopeptidase 2 (ERAP2) in complex with a hydroxamic derivative ligand | | 分子名称: | (2S)-N~1~-benzyl-2-[(4-fluorophenyl)methyl]-N~3~-hydroxypropanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Saridakis, E, Giastas, P, Mpakali, A, Deprez-Poulain, R, Stratikos, E. | | 登録日 | 2016-04-05 | | 公開日 | 2017-03-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
5K1V
 
 | | Crystal structure of Endoplasmic Reticulum aminopeptidase 2 (ERAP2) in complex with a diaminobenzoic acid derivative ligand. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | 著者 | Saridakis, E, Papakyriakou, A, Giastas, P, Mpakali, A, Mavridis, I.M, Stratikos, E. | | 登録日 | 2016-05-18 | | 公開日 | 2017-03-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.897 Å) | | 主引用文献 | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
