2JQM
 
 | | Yellow Fever Envelope Protein Domain III NMR Structure (S288-K398) | | 分子名称: | Envelope protein E | | 著者 | Volk, D.E, Gandham, S.H, May, F.J, Anderson, A, Barrett, A.D, Gorenstein, D.G. | | 登録日 | 2007-06-03 | | 公開日 | 2008-06-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of yellow fever virus envelope protein domain III.
Virology, 394, 2009
|
|
2JLN
 
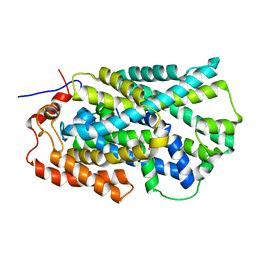 | | Structure of Mhp1, a nucleobase-cation-symport-1 family transporter | | 分子名称: | MERCURY (II) ION, MHP1, SODIUM ION | | 著者 | Weyand, S, Shimamura, T, Yajima, S, Suzuki, S, Mirza, O, Krusong, K, Carpenter, E.P, Rutherford, N.G, Hadden, J.M, O'Reilly, J, Ma, P, Saidijam, M, Patching, S.G, Hope, R.J, Norbertczak, H.T, Roach, P.C.J, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | 登録日 | 2008-09-11 | | 公開日 | 2008-10-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure and Molecular Mechanism of a Nucleobase-Cation-Symport-1 Family Transporter.
Science, 322, 2008
|
|
1QFG
 
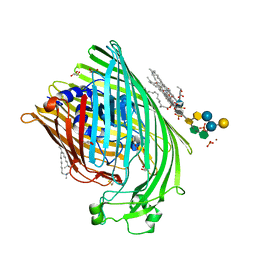 | | E. COLI FERRIC HYDROXAMATE RECEPTOR (FHUA) | | 分子名称: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | 著者 | Ferguson, A.D, Welte, W, Hofmann, E, Lindner, B, Holst, O, Coulton, J.W, Diederichs, K. | | 登録日 | 1999-04-10 | | 公開日 | 2000-07-26 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins.
Structure Fold.Des., 8, 2000
|
|
8C3E
 
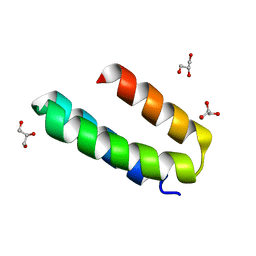 | | Engineered mini-protein LCB2 (blocking ligand of SARS-Cov-2 spike protein) | | 分子名称: | Engineered protein LCB2, GLYCEROL | | 著者 | Korban, S.A, Mikhailovskii, O.V, Luzik, D.A, Gurzhiy, V.V, Levkina, A.D, Kharkov, B.B, Skrynnikov, N.R. | | 登録日 | 2022-12-23 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Engineered mini-protein LCB2 (blocking ligand of SARS-Cov-2 spike protein)
To Be Published
|
|
1QJQ
 
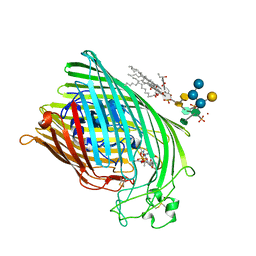 | | FERRIC HYDROXAMATE RECEPTOR FROM ESCHERICHIA COLI (FHUA) | | 分子名称: | 3-HYDROXY-TETRADECANOIC ACID, DIPHOSPHATE, FERRIC HYDROXAMATE RECEPTOR, ... | | 著者 | Ferguson, A.D, Braun, V, Fiedler, H.-P, Coulton, J.W, Diederichs, K, Welte, W. | | 登録日 | 1999-06-29 | | 公開日 | 2000-06-05 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA.
Protein Sci., 9, 2000
|
|
5BSY
 
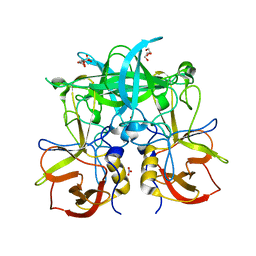 | |
2A02
 
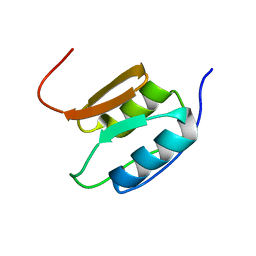 | | Solution NMR Structure of the Periplasmic Signaling Domain of the Outer Membrane Iron Transporter PupA from Pseudomonas putida. | | 分子名称: | Ferric-pseudobactin 358 receptor | | 著者 | Ferguson, A.D, Amezcua, C.A, Chelliah, Y, Rosen, M.K, Deisenhofer, J. | | 登録日 | 2005-06-15 | | 公開日 | 2006-09-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Signal transduction pathway of TonB-dependent transporters.
Proc.Natl.Acad.Sci.Usa, 2, 2006
|
|
1ZYR
 
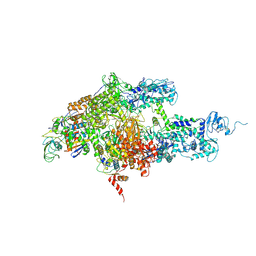 | | Structure of Thermus thermophilus RNA polymerase holoenzyme in complex with the antibiotic streptolydigin | | 分子名称: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase omega chain, ... | | 著者 | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | 登録日 | 2005-06-10 | | 公開日 | 2005-09-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation.
Cell(Cambridge,Mass.), 122, 2005
|
|
5BSX
 
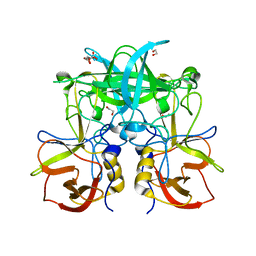 | |
1GSD
 
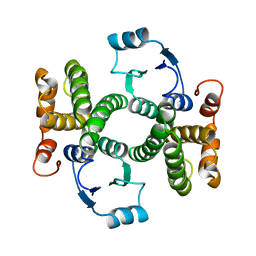 | |
1H4P
 
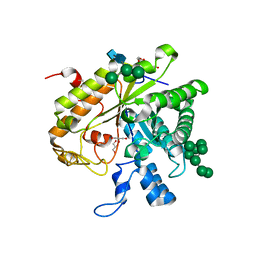 | | Crystal structure of exo-1,3-beta glucanse from Saccharomyces cerevisiae | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUCAN 1,3-BETA-GLUCOSIDASE I/II, GLYCEROL, ... | | 著者 | Ferguson, A.D. | | 登録日 | 2001-05-11 | | 公開日 | 2003-10-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The Er Protein Folding Sensor Udp-Glucose Glycoprotein:Glucosyltransferase Modifies Substrates Distant to Local Changes in Glycoprotein Conformation.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1GSE
 
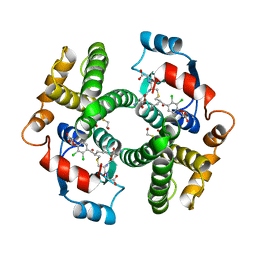 | |
8OV6
 
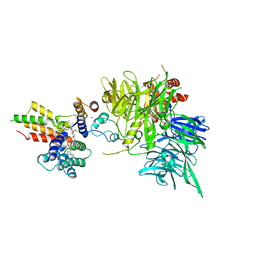 | | Ternary structure of intramolecular bivalent glue degrader IBG1 bound to BRD4 and DCAF16:DDB1deltaBPB | | 分子名称: | Bromodomain-containing protein 4, DDB1- and CUL4-associated factor 16, DDB1deltaBPB, ... | | 著者 | Cowan, A.D, Sundaramoorthy, R, Nakasone, M.A, Ciulli, A. | | 登録日 | 2023-04-25 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Targeted protein degradation via intramolecular bivalent glues.
Nature, 627, 2024
|
|
5TCZ
 
 | |
4UX6
 
 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, INDUCIBLE, ... | | 著者 | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | 登録日 | 2014-08-19 | | 公開日 | 2014-10-08 | | 最終更新日 | 2018-01-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4DMX
 
 | | Cathepsin K inhibitor | | 分子名称: | (1R,2R)-N-(1-cyanocyclopropyl)-2-{[4-(4-fluorophenyl)piperazin-1-yl]carbonyl}cyclohexanecarboxamide, Cathepsin K, GLYCEROL | | 著者 | Dossetter, A.G, Beeley, H, Bowyer, J, Cook, C.R, Crawford, J.J, Finlayson, J.E, Heron, N.M, Heyes, C, Highton, A.J, Hudson, J.A, Kenny, P.W, Martin, S, MacFaul, P.A, McGuire, T.M, Gutierrez, P.M, Morley, A.D, Morris, J.J, Page, K.M, Rosenbrier Ribeiro, L, Sawney, H, Steinbacher, S, Krapp, S, Jestel, A, Smith, C, Vickers, M. | | 登録日 | 2012-02-08 | | 公開日 | 2012-07-11 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | (1R,2R)-N-(1-cyanocyclopropyl)-2-(6-methoxy-1,3,4,5-tetrahydropyrido[4,3-b]indole-2-carbonyl)cyclohexanecarboxamide (AZD4996): a potent and highly selective cathepsin K inhibitor for the treatment of osteoarthritis.
J.Med.Chem., 55, 2012
|
|
4DMY
 
 | | Cathepsin K inhibitor | | 分子名称: | (1R,2R)-N-(1-cyanocyclopropyl)-2-[(8-fluoro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl)carbonyl]cyclohexanecarboxamide, Cathepsin K, GLYCEROL, ... | | 著者 | Dossetter, A.G, Beeley, H, Bowyer, J, Cook, C.R, Crawford, J.J, Finlayson, J.E, Heron, N.M, Heyes, C, Highton, A.J, Hudson, J.A, Kenny, P.W, Martin, S, MacFaul, P.A, McGuire, T.M, Gutierrez, P.M, Morley, A.D, Morris, J.J, Page, K.M, Rosenbrier Ribeiro, L, Sawney, H, Steinbacher, S, Krapp, S, Jestel, A, Smith, C, Vickers, M. | | 登録日 | 2012-02-08 | | 公開日 | 2012-07-11 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | (1R,2R)-N-(1-cyanocyclopropyl)-2-(6-methoxy-1,3,4,5-tetrahydropyrido[4,3-b]indole-2-carbonyl)cyclohexanecarboxamide (AZD4996): a potent and highly selective cathepsin K inhibitor for the treatment of osteoarthritis.
J.Med.Chem., 55, 2012
|
|
8OVB
 
 | |
8OSQ
 
 | |
8P9M
 
 | |
6W5H
 
 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5d | | 分子名称: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-03-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6W2A
 
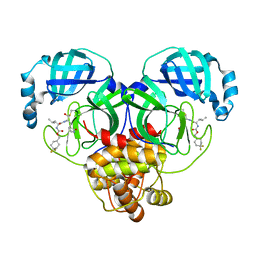 | | 1.65 A resolution structure of SARS-CoV 3CL protease in complex with inhibitor 7j | | 分子名称: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Replicase polyprotein 1a, [4,4-bis(fluoranyl)cyclohexyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | 著者 | Kashipathy, M.M, Lovell, S, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-03-05 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
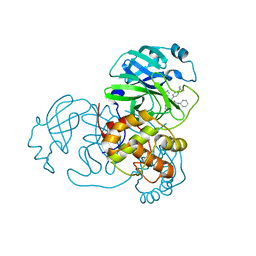
6W63
 
 | |
6W5J
 
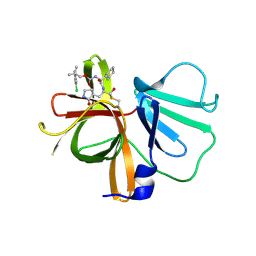 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7d | | 分子名称: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-03-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
2ZD1
 
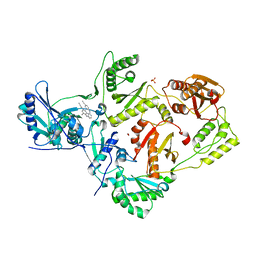 | | Crystal Structure of HIV-1 Reverse Transcriptase (RT) in Complex with TMC278 (Rilpivirine), A Non-nucleoside RT Inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, ... | | 著者 | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | 登録日 | 2007-11-16 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
