1EXM
 
 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS ELONGATION FACTOR TU (EF-TU) IN COMPLEX WITH THE GTP ANALOGUE GPPNHP. | | 分子名称: | ELONGATION FACTOR TU (EF-TU), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Hogg, T, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2000-05-03 | | 公開日 | 2000-06-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Insights into the GTPase Mechanism of EF-Tu from Structural Studies
The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions, 28, 2000
|
|
5IDK
 
 | |
4WUR
 
 | |
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | 分子名称: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-04-14 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | 分子名称: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2021-02-05 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
1DPW
 
 | | STRUCTURE OF HEN EGG-WHITE LYSOZYME IN COMPLEX WITH MPD | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Weiss, M.S, Palm, G.J, Hilgenfeld, R. | | 登録日 | 1999-12-28 | | 公開日 | 2000-01-03 | | 最終更新日 | 2011-11-23 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystallization, structure solution and refinement of hen egg-white lysozyme at pH 8.0 in the presence of MPD.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DG1
 
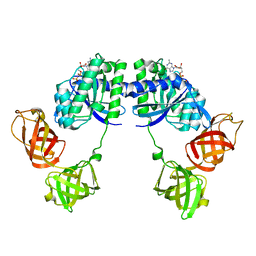 | | WHOLE, UNMODIFIED, EF-TU(ELONGATION FACTOR TU). | | 分子名称: | ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Abel, K, Yoder, M, Hilgenfeld, R, Jurnak, F. | | 登録日 | 1999-11-22 | | 公開日 | 1999-12-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | An alpha to beta conformational switch in EF-Tu.
Structure, 4, 1996
|
|
8R19
 
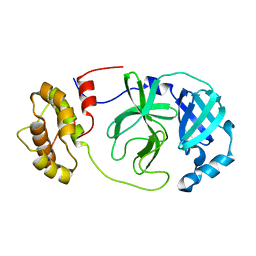 | |
8R0V
 
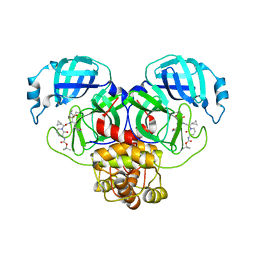 | |
8C9O
 
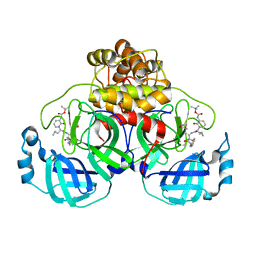 | |
8C9L
 
 | |
1HA3
 
 | | ELONGATION FACTOR TU IN COMPLEX WITH aurodox | | 分子名称: | BETA-MERCAPTOETHANOL, ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Vogeley, L, Palm, G.J, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2001-03-26 | | 公開日 | 2001-05-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Conformational Change of Elongation Factor TU Induced by Antibiotic Binding: Crystal Structure of the Complex between EF-TU:Gdp and Aurodox
J.Biol.Chem., 276, 2001
|
|
4YO1
 
 | |
4YNN
 
 | | Structure of Legionella pneumophila DegQ (S190A variant) | | 分子名称: | Hepta-peptide, Hexa-peptide, Octapeptide, ... | | 著者 | Wrase, R, Hilgenfeld, R, Hansen, G. | | 登録日 | 2015-03-10 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structures of DegQ from Legionella pneumophila Define Distinct ON and OFF States.
J.Mol.Biol., 427, 2015
|
|
7QUW
 
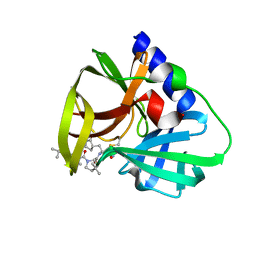 | | CVB3-3Cpro in complex with inhibitor MG-78 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, Protease 3C | | 著者 | Zhang, L, Hilgenfeld, R. | | 登録日 | 2022-01-19 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
7QUB
 
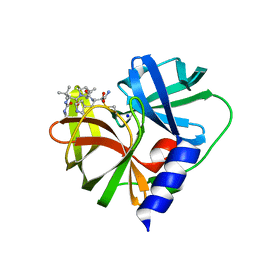 | | EV-A71-3Cpro in complex with inhibitor MG78 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, Protease 3C, SODIUM ION | | 著者 | El Kilani, H, Hilgenfeld, R. | | 登録日 | 2022-01-17 | | 公開日 | 2022-04-27 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
7QL8
 
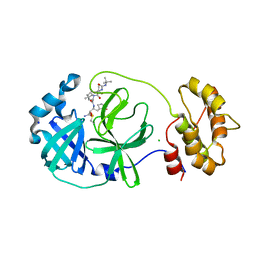 | | SARS-COV2 Main Protease in complex with inhibitor MG78 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | El Kilani, H, Hilgenfeld, R. | | 登録日 | 2021-12-19 | | 公開日 | 2022-04-27 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.807 Å) | | 主引用文献 | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
6Y2G
 
 | | Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | 分子名称: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | 登録日 | 2020-02-15 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
1F13
 
 | |
6FV1
 
 | | Structure of human coronavirus NL63 main protease in complex with the alpha-ketoamide (S)-N-((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)-2-cinnamamido-4-methylpentanamide (cinnamoyl-leucine-GlnLactam-CO-CO-NH-benzyl) | | 分子名称: | (2~{S})-4-methyl-~{N}-[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | 著者 | Zhang, L, Hilgenfeld, R. | | 登録日 | 2018-02-28 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication Structure-based design, synthesis, and activity assessment.
J.Med.Chem., 2020
|
|
6FV2
 
 | |
5HOL
 
 | |
5HIH
 
 | |
2VB0
 
 | | Crystal structure of coxsackievirus B3 proteinase 3C | | 分子名称: | CHLORIDE ION, POLYPROTEIN 3BCD | | 著者 | Anand, K, Mesters, J.R, Goerlach, R, Zell, R, Hilgenfeld, R. | | 登録日 | 2007-09-05 | | 公開日 | 2008-10-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of Coxsackie Virus B3 Proteinase 3C
To be Published
|
|
5LC0
 
 | | Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor | | 分子名称: | N-((S)-3-(4-(aminomethyl)phenyl)-1-(((R)-4-guanidino-1-(5-hydroxy-1,3,2-dioxaborinan-2-yl)butyl)amino)-1-oxopropan-2-yl)benzamide, NS2B-NS3 protease,NS2B-NS3 protease | | 著者 | Lei, J, Hansen, G, Zhang, L.L, Hilgenfeld, R. | | 登録日 | 2016-06-18 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor.
Science, 353, 2016
|
|
