1IPH
 
 | |
1BQU
 
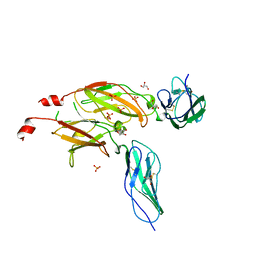 | | CYTOKYNE-BINDING REGION OF GP130 | | 分子名称: | GLYCEROL, PROTEIN (GP130), SULFATE ION | | 著者 | Bravo, J, Staunton, D, Heath, J.K, Jones, E.Y. | | 登録日 | 1998-08-18 | | 公開日 | 1998-08-26 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a cytokine-binding region of gp130.
EMBO J., 17, 1998
|
|
1H9D
 
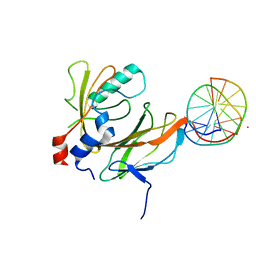 | | Aml1/cbf-beta/dna complex | | 分子名称: | CORE-BINDING FACTOR ALPHA SUBUNIT1, CORE-BINDING FACTOR CBF-BETA, DNA (5'-(*CP*AP*AP*CP*CP*GP*CP*AP*AP*C)-3'), ... | | 著者 | Bravo, J, Warren, A.J. | | 登録日 | 2001-03-07 | | 公開日 | 2001-03-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Leukemia-Associated Aml1 (Runx1)-Cbfbeta Complex Functions as a DNA-Induced Molecular Clamp
Nat.Struct.Biol., 8, 2001
|
|
2YDL
 
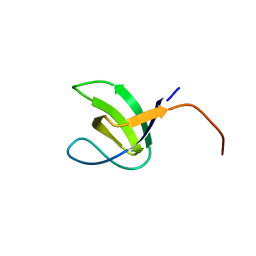 | | Crystal structure of SH3C from CIN85 | | 分子名称: | SH3 DOMAIN-CONTAINING KINASE-BINDING PROTEIN 1 | | 著者 | Bravo, J, Cardenes, N. | | 登録日 | 2011-03-22 | | 公開日 | 2012-03-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Distinct Ubiquitin Binding Modes Exhibited by SH3 Domains: Molecular Determinants and Functional Implications.
Plos One, 8, 2013
|
|
1H6H
 
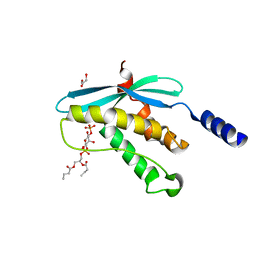 | | Structure of the PX domain from p40phox bound to phosphatidylinositol 3-phosphate | | 分子名称: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, GLYCEROL, NEUTROPHIL CYTOSOL FACTOR 4 | | 著者 | Karathanassis, D, Bravo, J, Pacold, M, Perisic, O, Williams, R.L. | | 登録日 | 2001-06-15 | | 公開日 | 2001-11-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Crystal Structure of the Px Domain from P40Phox Bound to Phosphatidylinositol 3-Phosphate
Mol.Cell, 8, 2001
|
|
4XOS
 
 | | ANP32A LRR domain | | 分子名称: | Acidic leucine-rich nuclear phosphoprotein 32 family member A, CHLORIDE ION, GLYCEROL | | 著者 | Zamora-Caballero, S, Bravo, J. | | 登録日 | 2015-01-16 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.559 Å) | | 主引用文献 | High-resolution crystal structure of the leucine-rich repeat domain of the human tumour suppressor PP32A (ANP32A).
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4AUV
 
 | | Crystal Structure of the BRMS1 N-terminal region | | 分子名称: | ACETIC ACID, BREAST CANCER METASTASIS SUPPRESSOR 1, CHLORIDE ION, ... | | 著者 | Spinola-Amilibia, M, Rivera, J, Ortiz-Lombardia, M, Romero, A, Neira, J.L, Bravo, J. | | 登録日 | 2012-05-22 | | 公開日 | 2013-03-20 | | 最終更新日 | 2019-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Brms151-98 and Brms151-84 are Crystal Oligomeric Coiled Coils with Different Oligomerization States, which Behave as Disordered Protein Fragments in Solution.
J.Mol.Biol., 425, 2013
|
|
2XUS
 
 | | Crystal Structure of the BRMS1 N-terminal region | | 分子名称: | BREAST CANCER METASTASIS-SUPPRESSOR 1, CHLORIDE ION, SULFATE ION | | 著者 | Spinola-Amilibia, M, Rivera, J, Ortiz-Lombardia, M, Romero, A, Neira, J.L, Bravo, J. | | 登録日 | 2010-10-20 | | 公開日 | 2011-07-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.912 Å) | | 主引用文献 | The Structure of Brms1 Nuclear Export Signal and Snx6 Interacting Region Reveals a Hexamer Formed by Antiparallel Coiled Coils.
J.Mol.Biol., 411, 2011
|
|
3CYJ
 
 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus | | 分子名称: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme-like protein, SODIUM ION | | 著者 | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Bravo, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-04-25 | | 公開日 | 2008-05-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus.
To be Published
|
|
4Z8B
 
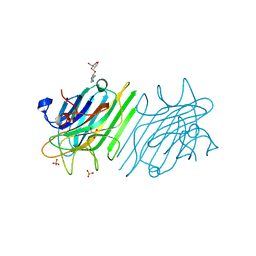 | | crystal structure of a DGL mutant - H51G H131N | | 分子名称: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, GLYCEROL, ... | | 著者 | Zamora-Caballero, S, Perez, A, Sanz, L, Bravo, J, Calvete, J.J. | | 登録日 | 2015-04-08 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | Quaternary structure of Dioclea grandiflora lectin assessed by equilibrium sedimentation and crystallographic analysis of recombinant mutants.
Febs Lett., 589, 2015
|
|
1GGE
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, NATIVE STRUCTURE AT 1.9 A RESOLUTION. | | 分子名称: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, PROTEIN (CATALASE HPII) | | 著者 | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | 登録日 | 2000-08-16 | | 公開日 | 2000-08-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
1GGJ
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, ASN201ALA VARIANT. | | 分子名称: | CATALASE HPII, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE | | 著者 | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | 登録日 | 2000-08-21 | | 公開日 | 2000-08-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
2AK5
 
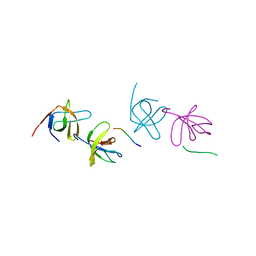 | | beta PIX-SH3 complexed with a Cbl-b peptide | | 分子名称: | 8-residue peptide from a signal transduction protein CBL-B, Rho guanine nucleotide exchange factor 7 | | 著者 | Jozic, D, Cardenes, N, Deribe, Y.L, Moncalian, G, Hoeller, D, Groemping, Y, Dikic, I, Rittinger, K, Bravo, J. | | 登録日 | 2005-08-03 | | 公開日 | 2005-10-11 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Cbl promotes clustering of endocytic adaptor proteins.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BZ8
 
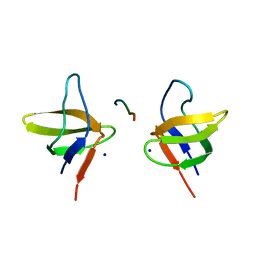 | | N-terminal Sh3 domain of CIN85 bound to Cbl-b peptide | | 分子名称: | SH3-DOMAIN KINASE BINDING PROTEIN 1, SIGNAL TRANSDUCTION PROTEIN CBL-B SH3-BINDING PROTEIN CBL-B, RING FINGER PROTEIN 56, ... | | 著者 | Cardenes, N, Moncalian, G, Bravo, J. | | 登録日 | 2005-08-12 | | 公開日 | 2005-10-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Cbl Promotes Clustering of Endocytic Adaptor Proteins
Nat.Struct.Mol.Biol., 12, 2005
|
|
1GGH
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, HIS128ALA VARIANT. | | 分子名称: | CATALASE HPII, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | 登録日 | 2000-08-21 | | 公開日 | 2000-08-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
1GGF
 
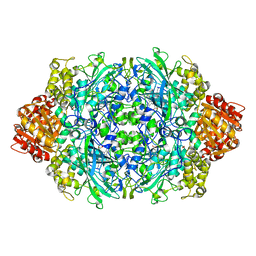 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, VARIANT HIS128ASN, COMPLEX WITH HYDROGEN PEROXIDE. | | 分子名称: | CATALASE HPII, HYDROGEN PEROXIDE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | 登録日 | 2000-08-21 | | 公開日 | 2000-08-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
1GGK
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, ASN201HIS VARIANT. | | 分子名称: | CATALASE HPII, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | 登録日 | 2000-08-21 | | 公開日 | 2000-08-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
4BMJ
 
 | | Structure of the UBZ1and2 tandem of the ubiquitin-binding adaptor protein TAX1BP1 | | 分子名称: | CHLORIDE ION, TAX1-BINDING PROTEIN 1, ZINC ION | | 著者 | Ceregido, M.A, Spinola-Amilibia, M, Buts, L, Rivera, J, Bravo, J, van Nuland, N.A.J. | | 登録日 | 2013-05-09 | | 公開日 | 2013-11-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The Structure of Tax1BP1 Ubz1 + 2 Provides Insight Into Target Specificity and Adaptability
J.Mol.Biol., 426, 2014
|
|
7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | 著者 | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | 登録日 | 2021-11-27 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDG
 
 | | SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | 登録日 | 2021-11-27 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
1GG9
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, HIS128ASN VARIANT. | | 分子名称: | CATALASE HPII, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | 登録日 | 2000-08-11 | | 公開日 | 2000-08-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
6EXZ
 
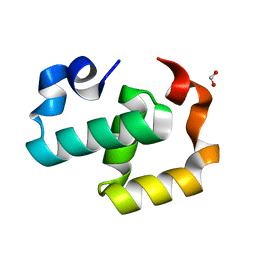 | | Crystal structure of Mex67 C-term | | 分子名称: | FORMIC ACID, mRNA export factor MEX67 | | 著者 | Mohamad, N, Bravo, J. | | 登録日 | 2017-11-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Mip6 binds directly to the Mex67 UBA domain to maintain low levels of Msn2/4 stress-dependent mRNAs.
Embo Rep., 2019
|
|
4U7A
 
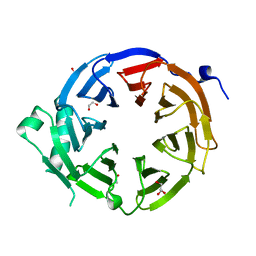 | |
1O7K
 
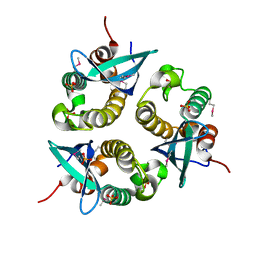 | | human p47 PX domain complex with sulphates | | 分子名称: | NEUTROPHIL CYTOSOL FACTOR 1, SULFATE ION | | 著者 | Karathanassis, D, Bravo, J, Perisic, O, Pacold, C.M, Williams, R.L. | | 登録日 | 2002-11-07 | | 公開日 | 2002-11-20 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Binding of the Px Domain of P47Phox to Phosphatidylinositol 3.4-Bisphosphate and Phosphatidic Acid is Masked by an Intramolecular Interaction
Embo J., 21, 2002
|
|
4A9A
 
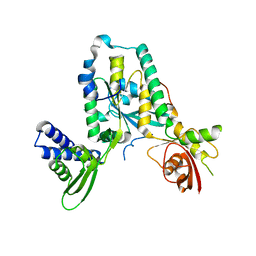 | | Structure of Rbg1 in complex with Tma46 dfrp domain | | 分子名称: | RIBOSOME-INTERACTING GTPASE 1, TRANSLATION MACHINERY-ASSOCIATED PROTEIN 46 | | 著者 | Francis, S.M, Gas, M, Daugeron, M, Seraphin, B, Bravo, J. | | 登録日 | 2011-11-25 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Rbg1-Tma46 Dimer Structure Reveals New Functional Domains and Their Role in Polysome Recruitment.
Nucleic Acids Res., 40, 2012
|
|
