6L0W
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | 分子名称: | 1,2-ETHANEDIOL, CITRATE ANION, NGR2, ... | | 著者 | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | 登録日 | 2019-09-27 | | 公開日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.591 Å) | | 主引用文献 | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
8KAD
 
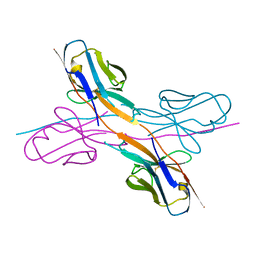 | | Crystal structure of an antibody light chain tetramer with 3D domain swapping | | 分子名称: | Antibody light chain | | 著者 | Sakai, T, Mashima, T, Kobayashi, N, Ogata, H, Uda, T, Hifumi, E, Hirota, S. | | 登録日 | 2023-08-02 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and thermodynamic insights into antibody light chain tetramer formation through 3D domain swapping.
Nat Commun, 14, 2023
|
|
5AUP
 
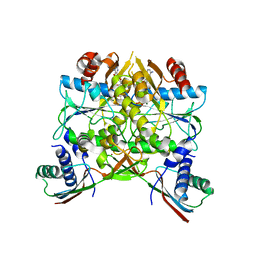 | | Crystal structure of the HypAB complex | | 分子名称: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | 著者 | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | 登録日 | 2015-05-27 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUN
 
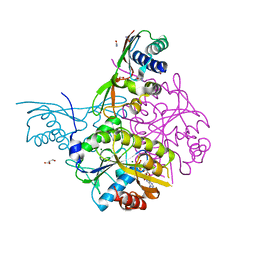 | | Crystal structure of the HypAB-Ni complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ParA/MinD family, ... | | 著者 | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | 登録日 | 2015-05-27 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1BDJ
 
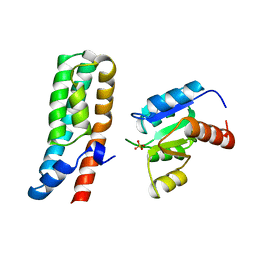 | | COMPLEX STRUCTURE OF HPT DOMAIN AND CHEY | | 分子名称: | AEROBIC RESPIRATION CONTROL SENSOR PROTEIN ARCB, CHEY, SULFATE ION | | 著者 | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | 登録日 | 1998-05-10 | | 公開日 | 1999-05-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor protein ArcB complexed with the chemotaxis response regulator CheY.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
8ZUH
 
 | | Crystal structure of bovine Fbs2/Skp1/Man3GlcNAc2 complex | | 分子名称: | F-box only protein 6, S-phase kinase-associated protein 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Satoh, T, Mizushima, T, Yagi, H, Kato, R, Kato, K. | | 登録日 | 2024-06-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis of sugar recognition by SCF FBS2 ubiquitin ligase involved in NGLY1 deficiency.
Febs Lett., 598, 2024
|
|
8HYE
 
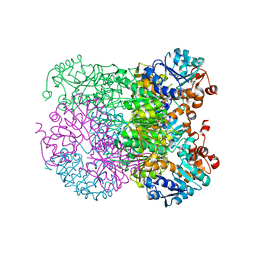 | | Structure of amino acid dehydrogenase-2752 with ligand | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alanine dehydrogenase, ... | | 著者 | Sakuraba, H, Ohshima, T. | | 登録日 | 2023-01-06 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Two different alanine dehydrogenases from Geobacillus kaustophilus: Their biochemical characteristics and differential expression in vegetative cells and spores.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
8HYH
 
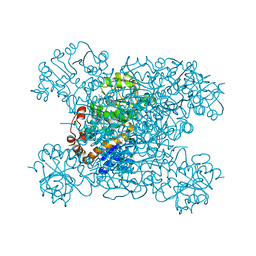 | | Structure of amino acid dehydrogenase3448 | | 分子名称: | 1,2-ETHANEDIOL, Alanine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Sakuraba, H, Ohshima, T. | | 登録日 | 2023-01-06 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Two different alanine dehydrogenases from Geobacillus kaustophilus: Their biochemical characteristics and differential expression in vegetative cells and spores.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
6KPD
 
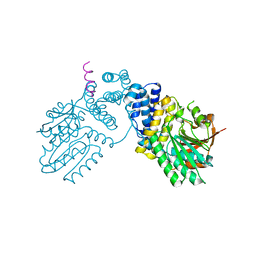 | |
6KPB
 
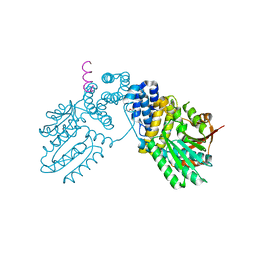 | |
5D94
 
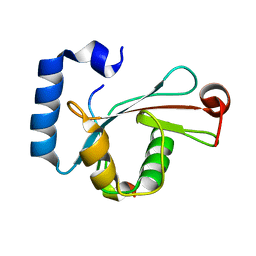 | | Crystal structure of LC3-LIR peptide complex | | 分子名称: | Microtubule-associated proteins 1A/1B light chain 3B, Peptide from FYVE and coiled-coil domain-containing protein 1 | | 著者 | Takagi, K, Mizushima, T, Johansen, T. | | 登録日 | 2015-08-18 | | 公開日 | 2015-10-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | FYCO1 Contains a C-terminally Extended, LC3A/B-preferring LC3-interacting Region (LIR) Motif Required for Efficient Maturation of Autophagosomes during Basal Autophagy
J.Biol.Chem., 290, 2015
|
|
7WDT
 
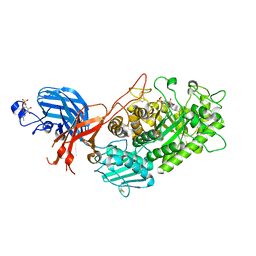 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with GlcNAc-6S | | 分子名称: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ... | | 著者 | Yamada, C, Kashima, T, Fushinobu, S, Katoh, T, Katayama, T. | | 登録日 | 2021-12-22 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
1IPA
 
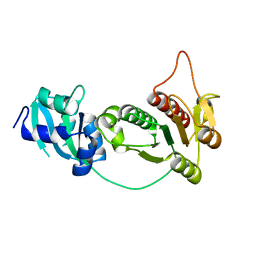 | | CRYSTAL STRUCTURE OF RNA 2'-O RIBOSE METHYLTRANSFERASE | | 分子名称: | RNA 2'-O-RIBOSE METHYLTRANSFERASE | | 著者 | Nureki, O, Shirouzu, M, Hashimoto, K, Ishitani, R, Terada, T, Tamakoshi, M, Oshima, T, Chijimatsu, M, Takio, K, Vassylyev, D.G, Shibata, T, Inoue, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2001-05-02 | | 公開日 | 2002-07-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | An enzyme with a deep trefoil knot for the active-site architecture.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7BQS
 
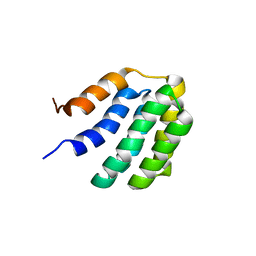 | | Solution NMR structure of fold-U Nomur; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Nomur | | 著者 | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQR
 
 | | Solution NMR structure of fold-K Mussoc; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Mussoc | | 著者 | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
1A2B
 
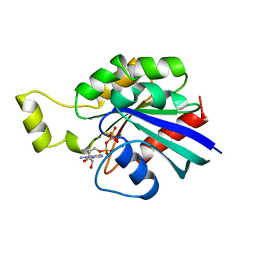 | | HUMAN RHOA COMPLEXED WITH GTP ANALOGUE | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, TRANSFORMING PROTEIN RHOA | | 著者 | Ihara, K, Muraguchi, S, Kato, M, Shimizu, T, Shirakawa, M, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | 登録日 | 1997-12-26 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue.
J.Biol.Chem., 273, 1998
|
|
7DFP
 
 | | Human dopamine D2 receptor in complex with spiperone | | 分子名称: | 8-[4-(4-fluorophenyl)-4-oxidanylidene-butyl]-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, D(2) dopamine receptor,Soluble cytochrome b562, FabH, ... | | 著者 | Im, D, Shimamura, T, Iwata, S. | | 登録日 | 2020-11-09 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of the dopamine D 2 receptor in complex with the antipsychotic drug spiperone.
Nat Commun, 11, 2020
|
|
1AUG
 
 | | CRYSTAL STRUCTURE OF THE PYROGLUTAMYL PEPTIDASE I FROM BACILLUS AMYLOLIQUEFACIENS | | 分子名称: | PYROGLUTAMYL PEPTIDASE-1 | | 著者 | Odagaki, Y, Hayashi, A, Okada, K, Hirotsu, K, Kabashima, T, Ito, K, Yoshimoto, T, Tsuru, D, Sato, M, Clardy, J. | | 登録日 | 1997-08-26 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of pyroglutamyl peptidase I from Bacillus amyloliquefaciens reveals a new structure for a cysteine protease.
Structure Fold.Des., 7, 1999
|
|
2Z7C
 
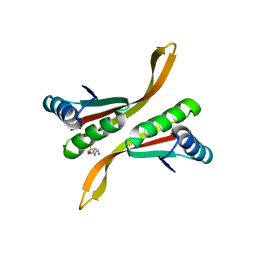 | | Crystal structure of chromatin protein alba from hyperthermophilic archaeon pyrococcus horikoshii | | 分子名称: | ARGININE, DNA/RNA-binding protein Alba | | 著者 | Hada, K, Nakashima, T, Osawa, T, Shimada, H, Kakuta, Y, Kimura, M. | | 登録日 | 2007-08-17 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure and functional analysis of an archaeal chromatin protein Alba from the hyperthermophilic archaeon Pyrococcus horikoshii OT3.
Biosci.Biotechnol.Biochem., 72, 2008
|
|
5B3G
 
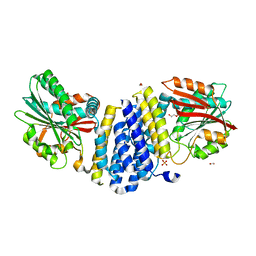 | | The crystal structure of the heterodimer of SHORT-ROOT and SCARECROW GRAS domains | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Hirano, Y, Nakagawa, M, Hakoshima, T. | | 登録日 | 2016-02-29 | | 公開日 | 2017-03-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD
Nat Plants, 3, 2017
|
|
5B3H
 
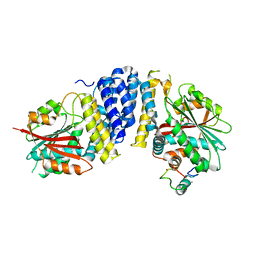 | | The crystal structure of the JACKDAW/IDD10 bound to the heterodimeric SHR-SCR complex | | 分子名称: | Protein SCARECROW, Protein SHORT-ROOT, ZINC ION, ... | | 著者 | Hirano, Y, Suyama, T, Nakagawa, M, Hakoshima, T. | | 登録日 | 2016-02-29 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD
Nat Plants, 3, 2017
|
|
5B5W
 
 | | Crystal structure of MOB1-LATS1 NTR domain complex | | 分子名称: | MOB kinase activator 1B, Serine/threonine-protein kinase LATS1, ZINC ION | | 著者 | KIM, S.-Y, Tachioka, Y, Mori, T, Hakoshima, T. | | 登録日 | 2016-05-24 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.957 Å) | | 主引用文献 | Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Sci Rep, 6, 2016
|
|
5B6B
 
 | | Complex of LATS1 and phosphomimetic MOB1b | | 分子名称: | CHLORIDE ION, MOB kinase activator 1B, Serine/threonine-protein kinase LATS1, ... | | 著者 | KIM, S.-Y, Tachioka, Y, Mori, T, Hakoshima, T. | | 登録日 | 2016-05-26 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.536 Å) | | 主引用文献 | Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Sci Rep, 6, 2016
|
|
7BQC
 
 | | Solution NMR structure of NF4; de novo designed protein with a novel fold | | 分子名称: | NF4 | | 著者 | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, T, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
2H9V
 
 | | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y27632 | | 分子名称: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, Rho-associated protein kinase 2 | | 著者 | Yamaguchi, H, Miwa, Y, Kasa, M, Kitano, K, Amano, M, Kaibuchi, K, Hakoshima, T. | | 登録日 | 2006-06-12 | | 公開日 | 2006-12-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y-27632
J.Biochem.(Tokyo), 140, 2006
|
|
