7VR7
 
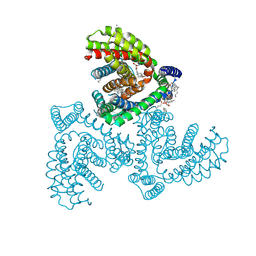 | | Inward-facing structure of human EAAT2 in the WAY213613-bound state | | 分子名称: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2021-10-22 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
8Y0H
 
 | | Structure of CXCR3 in complex with VUF11418 (Receptor-ligand focused map) | | 分子名称: | C-X-C chemokine receptor type 3, [(1~{R},5~{S})-6,6-dimethyl-2-bicyclo[3.1.1]hept-2-enyl]methyl-[[4-(2-iodanylphenyl)phenyl]methyl]-dimethyl-azanium | | 著者 | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | 登録日 | 2024-01-22 | | 公開日 | 2025-02-26 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Structural visualization of small molecule recognition by CXCR3 uncovers dual-agonism in the CXCR3-CXCR7 system.
Nat Commun, 16, 2025
|
|
8Y0N
 
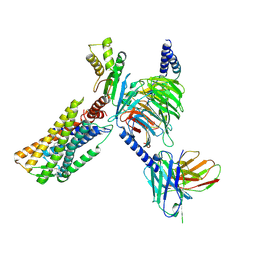 | | Structure of CXCR3 in complex with VUF11418 and Go (Full map) | | 分子名称: | Antibody fragment ScFv16, C-X-C chemokine receptor type 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | 登録日 | 2024-01-22 | | 公開日 | 2025-02-26 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structural visualization of small molecule recognition by CXCR3 uncovers dual-agonism in the CXCR3-CXCR7 system.
Nat Commun, 16, 2025
|
|
8XYK
 
 | | Structure of CXCR3 in complex with VUF10661 and Go (Full map) | | 分子名称: | (3~{S})-~{N}-[(2~{S})-6-azanyl-1-(2,2-diphenylethylamino)-1-oxidanylidene-hexan-2-yl]-2-(4-oxidanylidene-4-phenyl-butanoyl)-3,4-dihydro-1~{H}-isoquinoline-3-carboxamide, Antibody fragment ScFv16, C-X-C chemokine receptor type 3, ... | | 著者 | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | 登録日 | 2024-01-19 | | 公開日 | 2025-02-26 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Structural visualization of small molecule recognition by CXCR3 uncovers dual-agonism in the CXCR3-CXCR7 system.
Nat Commun, 16, 2025
|
|
8XXY
 
 | | Structure of CXCR3 in the apo-state (Receptor focused map) | | 分子名称: | C-X-C chemokine receptor type 3 | | 著者 | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | 登録日 | 2024-01-19 | | 公開日 | 2025-02-26 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Structural visualization of small molecule recognition by CXCR3 uncovers dual-agonism in the CXCR3-CXCR7 system.
Nat Commun, 16, 2025
|
|
8XYI
 
 | | Structure of CXCR3 in complex with VUF10661 (Receptor-ligand focused map) | | 分子名称: | (3~{S})-~{N}-[(2~{S})-6-azanyl-1-(2,2-diphenylethylamino)-1-oxidanylidene-hexan-2-yl]-2-(4-oxidanylidene-4-phenyl-butanoyl)-3,4-dihydro-1~{H}-isoquinoline-3-carboxamide, C-X-C chemokine receptor type 3 | | 著者 | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | 登録日 | 2024-01-19 | | 公開日 | 2025-02-26 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Structural visualization of small molecule recognition by CXCR3 uncovers dual-agonism in the CXCR3-CXCR7 system.
Nat Commun, 16, 2025
|
|
8XXZ
 
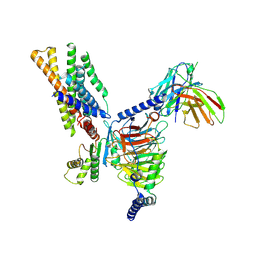 | | Structure of CXCR3 in the apo-state (Full map) | | 分子名称: | Antibody fragment ScFv16, C-X-C chemokine receptor type 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Sano, F.K, Saha, S, Sharma, S, Ganguly, M, Shihoya, W, Nureki, O, Shukla, A.K, Banerjee, R. | | 登録日 | 2024-01-19 | | 公開日 | 2025-02-26 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural visualization of small molecule recognition by CXCR3 uncovers dual-agonism in the CXCR3-CXCR7 system.
Nat Commun, 16, 2025
|
|
5HRT
 
 | | Crystal structure of mouse autotaxin in complex with a DNA aptamer | | 分子名称: | CALCIUM ION, CHLORIDE ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | 著者 | Kato, K, Nishimasu, H, Morita, J, Ishitani, R, Nureki, O. | | 登録日 | 2016-01-24 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | Structural basis for specific inhibition of Autotaxin by a DNA aptamer
Nat.Struct.Mol.Biol., 23, 2016
|
|
8JF0
 
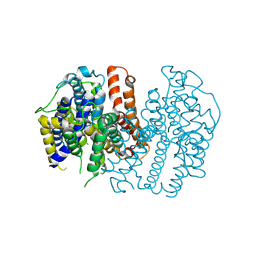 | |
8JF1
 
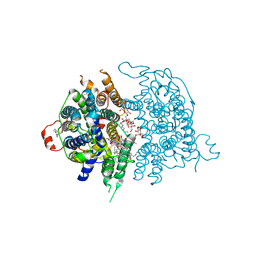 | | Human sodium-dependent vitamin C transporter 1 in an apo occluded state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Kobayashi, T.A, Kusakizako, T, Nureki, O. | | 登録日 | 2023-05-16 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-12-04 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Dimeric transport mechanism of human vitamin C transporter SVCT1.
Nat Commun, 15, 2024
|
|
8YGJ
 
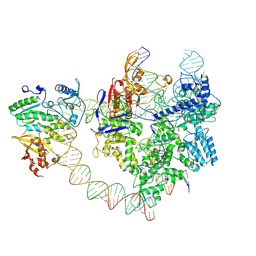 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 28-nt) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2024-02-26 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8JEZ
 
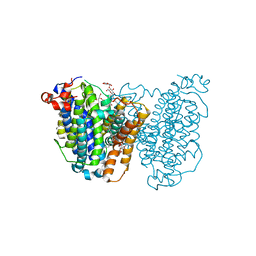 | | Human sodium-dependent vitamin C transporter 1 in an apo inward-open state | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Kobayashi, T.A, Kusakizako, T, Nureki, O. | | 登録日 | 2023-05-16 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-12-04 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Dimeric transport mechanism of human vitamin C transporter SVCT1.
Nat Commun, 15, 2024
|
|
8YNY
 
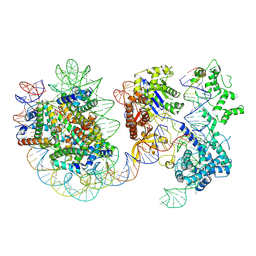 | | Structure of Cas9-sgRNA ribonucleoprotein bound to nucleosome | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (17-mer), DNA (175-mer), ... | | 著者 | Nagamura, R, Kujirai, T, Kusakizako, T, Hirano, H, Kurumizaka, H, Nureki, O. | | 登録日 | 2024-03-12 | | 公開日 | 2025-01-22 | | 実験手法 | ELECTRON MICROSCOPY (4.52 Å) | | 主引用文献 | Structural insights into how Cas9 targets nucleosomes.
Nat Commun, 15, 2024
|
|
8YH6
 
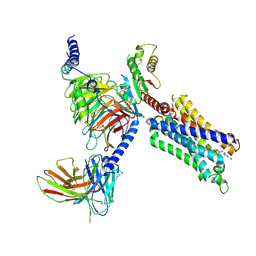 | | A3R-Gi complex bound to namodenoson | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2,Guanine nucleotide-binding protein G(i) subunit alpha-1 chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Hemagglutinin,Adenosine receptor A3,GFP chimera, ... | | 著者 | Oshima, H.S, Shihoya, W, Nureki, O. | | 登録日 | 2024-02-27 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Structural insights into the agonist selectivity of the adenosine A 3 receptor.
Nat Commun, 15, 2024
|
|
8YH0
 
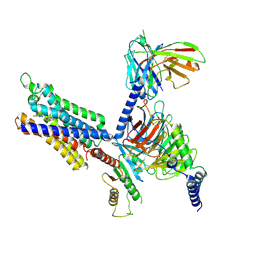 | | A3R-Gi complex bound to NECA | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2,Guanine nucleotide-binding protein G(i) subunit alpha-1, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, HA tag,Adenosine receptor A3,LgBiT,eGFP chimera, ... | | 著者 | Oshima, H.S, Shihoya, W, Nureki, O. | | 登録日 | 2024-02-27 | | 公開日 | 2024-11-06 | | 最終更新日 | 2024-11-27 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structural insights into the agonist selectivity of the adenosine A 3 receptor.
Nat Commun, 15, 2024
|
|
8YH5
 
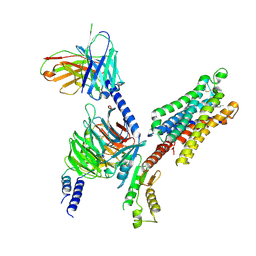 | | A3R-Gi complex bound to i6A | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2,Guanine nucleotide-binding protein G(i) subunit alpha-1 chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Hemagglutinin,Adenosine receptor A3,GFP chimera, ... | | 著者 | Oshima, H.S, Shihoya, W, Nureki, O. | | 登録日 | 2024-02-27 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Structural insights into the agonist selectivity of the adenosine A 3 receptor.
Nat Commun, 15, 2024
|
|
8YH2
 
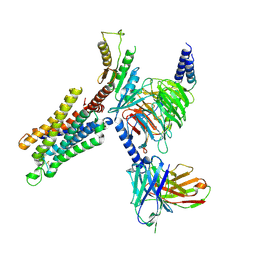 | | A3R-Gi complex bound to adenosine | | 分子名称: | ADENOSINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2,Guanine nucleotide-binding protein G(i) subunit alpha-1 chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Oshima, H.S, Shihoya, W, Nureki, O. | | 登録日 | 2024-02-27 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Structural insights into the agonist selectivity of the adenosine A 3 receptor.
Nat Commun, 15, 2024
|
|
8YH3
 
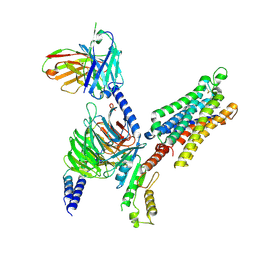 | | A3R-Gi complex bound to m6A | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2,Guanine nucleotide-binding protein G(i) subunit alpha-1 chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Hemagglutinin,Adenosine receptor A3,GFP chimera, ... | | 著者 | Oshima, H.S, Shihoya, W, Nureki, O. | | 登録日 | 2024-02-27 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural insights into the agonist selectivity of the adenosine A 3 receptor.
Nat Commun, 15, 2024
|
|
8GUJ
 
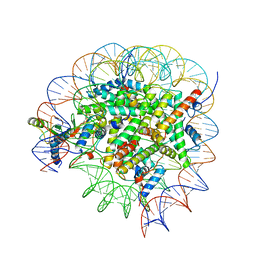 | | Bre1-nucleosome complex (Model II) | | 分子名称: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | 著者 | Onishi, S, Sato, K, Hamada, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | 登録日 | 2022-09-12 | | 公開日 | 2023-09-20 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
5I20
 
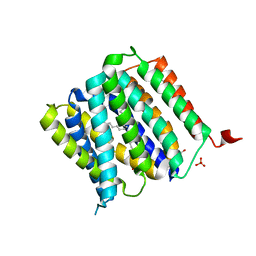 | | Crystal structure of protein | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Uncharacterized protein | | 著者 | Ishitani, R, Nureki, O. | | 登録日 | 2016-02-08 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for amino acid export by DMT superfamily transporter YddG.
Nature, 534, 2016
|
|
8GUI
 
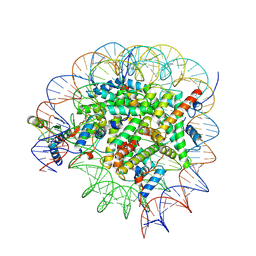 | | Bre1-nucleosome complex (Model I) | | 分子名称: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | 著者 | Onishi, S, Hamada, K, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | 登録日 | 2022-09-12 | | 公開日 | 2023-09-20 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
8GUK
 
 | | Human nucleosome core particle (free form) | | 分子名称: | DNA (147-mer), Histone H2A type 1, Histone H2B type 1-J, ... | | 著者 | Onishi, S, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | 登録日 | 2022-09-12 | | 公開日 | 2023-09-20 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
8JAF
 
 | | Structure of Muscarinic receptor (M2R) in complex with beta-arrestin1 (Local Refine, non-cross linked) | | 分子名称: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | 著者 | Maharana, J, Sano, F.K, Shihoya, W, Banerjee, R, Nureki, O, Shukla, A.K. | | 登録日 | 2023-05-05 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8HB0
 
 | | Structure of human SGLT2-MAP17 complex with TA1887 | | 分子名称: | (2R,3R,4S,5S,6R)-2-[3-[(4-cyclopropylphenyl)methyl]-4-fluoranyl-indol-1-yl]-6-(hydroxymethyl)oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, PDZK1-interacting protein 1, ... | | 著者 | Hiraizumi, M, Kishida, H, Miyaguchi, I, Nureki, O. | | 登録日 | 2022-10-27 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HG7
 
 | | Structure of human SGLT2-MAP17 complex with Sotagliflozin | | 分子名称: | (2S,3R,4R,5S,6R)-2-[4-chloranyl-3-[(4-ethoxyphenyl)methyl]phenyl]-6-methylsulfanyl-oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, PDZK1-interacting protein 1, ... | | 著者 | Hiraizumi, M, Kishida, H, Miyaguchi, I, Nureki, O. | | 登録日 | 2022-11-14 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
