3HJD
 
 | |
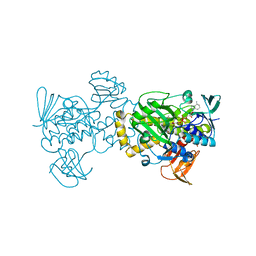
5XGV
 
 | |
4YO8
 
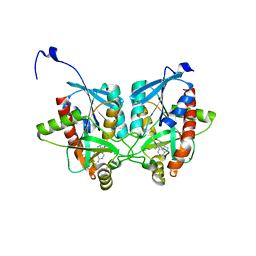 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)(hexyl)amino)methanol | | 分子名称: | Aminodeoxyfutalosine nucleosidase, ZINC ION, {[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl](hexyl)amino}methanol | | 著者 | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | 登録日 | 2015-03-11 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4YNB
 
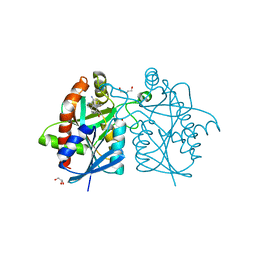 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with pyrazinylthio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(pyrazin-2-ylsulfanyl)methyl]pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | 登録日 | 2015-03-09 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
8IJD
 
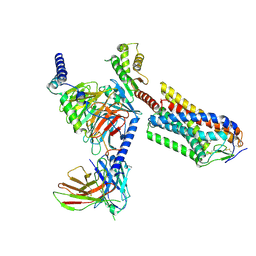 | | Cryo-EM structure of human HCAR2-Gi complex with MK-6892 | | 分子名称: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Pan, X, Fang, Y. | | 登録日 | 2023-02-27 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJ3
 
 | | Cryo-EM structure of human HCAR2-Gi complex without ligand (apo state) | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Pan, X, Fang, Y. | | 登録日 | 2023-02-24 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJB
 
 | | Cryo-EM structure of human HCAR2-Gi complex with acipimox | | 分子名称: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Pan, X, Fang, Y. | | 登録日 | 2023-02-27 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJA
 
 | | Cryo-EM structure of human HCAR2-Gi complex with niacin | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Pan, X, Fang, Y. | | 登録日 | 2023-02-26 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (2.69 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
7FCQ
 
 | |
7FCP
 
 | |
6JK8
 
 | | Cryo-EM structure of the full-length human IGF-1R in complex with insulin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, ... | | 著者 | Zhang, X, Yu, D, Wang, T. | | 登録日 | 2019-02-27 | | 公開日 | 2020-03-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Visualization of Ligand-Bound Ectodomain Assembly in the Full-Length Human IGF-1 Receptor by Cryo-EM Single-Particle Analysis.
Structure, 28, 2020
|
|
6MSJ
 
 | |
6MSG
 
 | |
6MSE
 
 | |
6MSB
 
 | |
6MSK
 
 | |
6MSH
 
 | |
5EAK
 
 | |
6MSD
 
 | |
3SLU
 
 | | Crystal structure of NMB0315 | | 分子名称: | M23 peptidase domain protein, NICKEL (II) ION | | 著者 | Shen, Y, Wang, X, Yang, X, Xu, H. | | 登録日 | 2011-06-26 | | 公開日 | 2012-02-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal structure of outer membrane protein NMB0315 from Neisseria meningitidis.
Plos One, 6, 2011
|
|
3STO
 
 | | Serpin from the trematode Schistosoma Haematobium | | 分子名称: | Serine protease inhibitor | | 著者 | Granzin, J, Weiergraeber, O.H, Lee, X, Blanton, R.E. | | 登録日 | 2011-07-11 | | 公開日 | 2012-05-30 | | 最終更新日 | 2013-01-23 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Three-dimensional structure of a schistosome serpin revealing an unusual configuration of the helical subdomain.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6AVL
 
 | | Orthorhombic Trypsin (295 K) in the presence of 50% xylose | | 分子名称: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | 著者 | Juers, D.H. | | 登録日 | 2017-09-02 | | 公開日 | 2017-09-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6B6R
 
 | |
6B6Q
 
 | |
6B6N
 
 | | Orthorhombic trypsin (295 K) in the presence of 50% mpd | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, BENZAMIDINE, CALCIUM ION, ... | | 著者 | Juers, D.H. | | 登録日 | 2017-10-03 | | 公開日 | 2017-11-01 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
