8CXN
 
 | |
8CY9
 
 | |
8CY7
 
 | |
7RTC
 
 | |
7ZC4
 
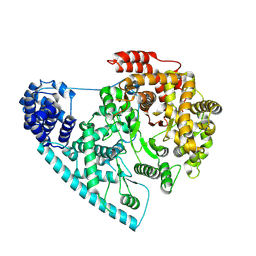 | | Cryo-EM structure of POLRMT mutant. | | 分子名称: | DNA-directed RNA polymerase, mitochondrial | | 著者 | Das, H, Hallberg, B.M. | | 登録日 | 2022-03-25 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Non-coding 7S RNA inhibits transcription via mitochondrial RNA polymerase dimerization.
Cell, 185, 2022
|
|
3JCO
 
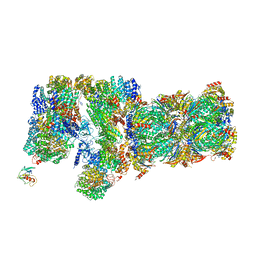 | | Structure of yeast 26S proteasome in M1 state derived from Titan dataset | | 分子名称: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | 著者 | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | 登録日 | 2016-01-06 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6IEA
 
 | | Structure of RVFV Gn and human monoclonal antibody R13 | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NSmGnGc, ... | | 著者 | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | 登録日 | 2018-09-13 | | 公開日 | 2019-04-10 | | 最終更新日 | 2019-07-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
6IEB
 
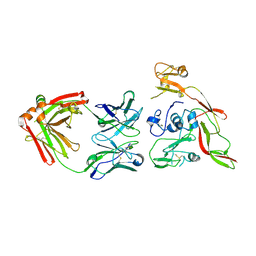 | | Structure of RVFV Gn and human monoclonal antibody R15 | | 分子名称: | NSmGnGc, R15 H chain, R15 L chain | | 著者 | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | 登録日 | 2018-09-13 | | 公開日 | 2019-04-10 | | 最終更新日 | 2019-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.409 Å) | | 主引用文献 | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
7N85
 
 | | Inner ring spoke from the isolated yeast NPC | | 分子名称: | Nucleoporin ASM4, Nucleoporin NIC96, Nucleoporin NSP1, ... | | 著者 | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | 登録日 | 2021-06-13 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N84
 
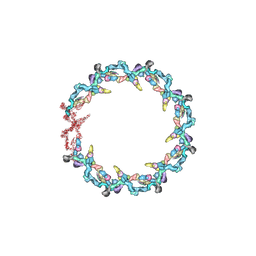 | | Double nuclear outer ring from the isolated yeast NPC | | 分子名称: | Nucleoporin 145c, Nucleoporin NUP120, Nucleoporin NUP133, ... | | 著者 | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | 登録日 | 2021-06-13 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (11.6 Å) | | 主引用文献 | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | 分子名称: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | 著者 | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | 登録日 | 2021-06-17 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (37 Å) | | 主引用文献 | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
3RER
 
 | | Crystal structure of E. coli Hfq in complex with AU6A RNA and ADP | | 分子名称: | 5'-R(*AP*UP*UP*UP*UP*UP*UP*A)-3', ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wang, W.W, Wu, J.H, Shi, Y.Y. | | 登録日 | 2011-04-05 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Cooperation of Escherichia coli Hfq hexamers in DsrA binding.
Genes Dev., 25, 2011
|
|
6M6C
 
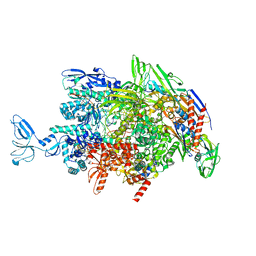 | | CryoEM structure of Thermus thermophilus RNA polymerase elongation complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Shi, J, Wen, A, Feng, Y. | | 登録日 | 2020-03-14 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6IEC
 
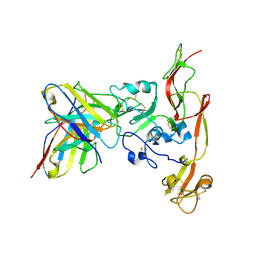 | | Structure of RVFV Gn and human monoclonal antibody R17 | | 分子名称: | NSmGnGc, R17 H chain, R17 L chain | | 著者 | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | 登録日 | 2018-09-13 | | 公開日 | 2019-04-10 | | 最終更新日 | 2019-07-10 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
6M6A
 
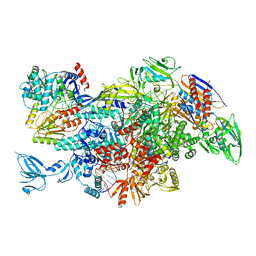 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Shi, J, Wen, A, Feng, Y. | | 登録日 | 2020-03-14 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6CF0
 
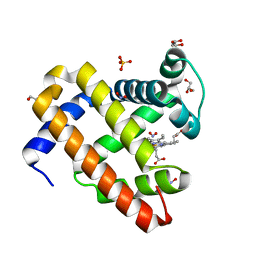 | | Sperm Whale Myoglobin H64V Mutant with Nitrite | | 分子名称: | GLYCEROL, Myoglobin, NITRITE ION, ... | | 著者 | Powell, S.M, Wang, B, Thomas, L.M, Richter-Addo, G.B. | | 登録日 | 2018-02-13 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
6M6B
 
 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase and ATP-gamma-S | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Shi, J, Wen, A, Feng, Y. | | 登録日 | 2020-03-14 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6IEK
 
 | | Structure of RVFV Gn and human monoclonal antibody R12 | | 分子名称: | Heavy chain of Fab R12, Light chain of Fab R12, NSmGnGc | | 著者 | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | 登録日 | 2018-09-14 | | 公開日 | 2019-04-10 | | 最終更新日 | 2019-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
3RES
 
 | |
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | 分子名称: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | 著者 | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | 登録日 | 2016-12-19 | | 公開日 | 2017-01-18 | | 最終更新日 | 2020-10-07 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
5MPS
 
 | | Structure of a spliceosome remodeled for exon ligation | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | 登録日 | 2016-12-18 | | 公開日 | 2017-01-18 | | 最終更新日 | 2020-10-07 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
4QWP
 
 | | co-crystal structure of chitosanase OU01 with substrate | | 分子名称: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Lyu, Q, Liu, W, Han, B. | | 登録日 | 2014-07-17 | | 公開日 | 2015-07-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and biochemical insights into the degradation mechanism of chitosan by chitosanase OU01.
Biochim.Biophys.Acta, 1850, 2015
|
|
5CLB
 
 | |
5CL9
 
 | |
5CL7
 
 | |
