3HUD
 
 | | THE STRUCTURE OF HUMAN BETA 1 BETA 1 ALCOHOL DEHYDROGENASE: CATALYTIC EFFECTS OF NON-ACTIVE-SITE SUBSTITUTIONS | | 分子名称: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | 著者 | Hurley, T.D, Bosron, W.F, Hamilton, J.A, Amzel, L.M. | | 登録日 | 1993-01-04 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of human beta 1 beta 1 alcohol dehydrogenase: catalytic effects of non-active-site substitutions.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
3I0N
 
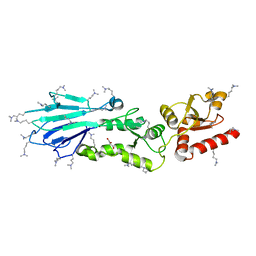 | | Structure of the S. pombe Nbs1 FHA/BRCT-repeat domain | | 分子名称: | DNA repair and telomere maintenance protein nbs1, GLYCEROL | | 著者 | Clapperton, J.A, Lloyd, J, Chapman, J.R, Jackson, S.P, Smerdon, S.J. | | 登録日 | 2009-06-25 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage
Cell(Cambridge,Mass.), 139, 2009
|
|
5G1M
 
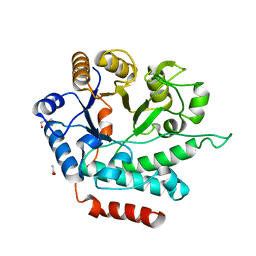 | | Crystal structure of NagZ from Pseudomonas aeruginosa | | 分子名称: | ACETATE ION, BETA-HEXOSAMINIDASE, CHLORIDE ION, ... | | 著者 | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | 登録日 | 2016-03-28 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5G22
 
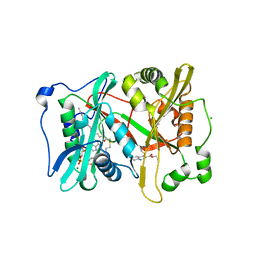 | | Plasmodium vivax N-myristoyltransferase in complex with a quinoline inhibitor (compound 26) | | 分子名称: | 2-oxopentadecyl-CoA, CHLORIDE ION, ETHYL 4-[(2-CYANOETHYL)SULFANYL]-6-{[6-(PIPERAZIN-1-YL), ... | | 著者 | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | 登録日 | 2016-04-06 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
5G20
 
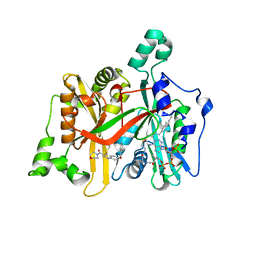 | | Leishmania major N-myristoyltransferase in complex with a quinoline inhibitor (compound 19). | | 分子名称: | 6-(BENZYLOXY)-4-(ETHYLSULFANYL)-3-[(MORPHOLIN-4-YL), DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | 著者 | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | 登録日 | 2016-04-06 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
5G3O
 
 | | Bacillus cereus formamidase (BceAmiF) inhibited with urea. | | 分子名称: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, FORMAMIDASE, ... | | 著者 | Gavira, J.A, Martinez-Rodriguez, S, Conejero-Muriel, M. | | 登録日 | 2016-04-29 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A novel cysteine carbamoyl-switch is responsible for the inhibition of formamidase, a nitrilase superfamily member.
Arch.Biochem.Biophys., 662, 2019
|
|
5G2M
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-HEXOSAMINIDASE | | 著者 | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | 登録日 | 2016-04-09 | | 公開日 | 2017-05-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5GNB
 
 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 2.3 angstrom, native-SAD phasing) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | 登録日 | 2016-07-20 | | 公開日 | 2017-06-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
5GAI
 
 | | Probabilistic Structural Models of Mature P22 Bacteriophage Portal, Hub, and Tailspike proteins | | 分子名称: | Peptidoglycan hydrolase gp4, Portal protein, Tail fiber protein | | 著者 | Pintilie, G, Chen, D.H, Haase-Pettingell, C.A, King, J.A, Chiu, W. | | 登録日 | 2015-12-01 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (10.5 Å) | | 主引用文献 | Resolution and Probabilistic Models of Components in CryoEM Maps of Mature P22 Bacteriophage.
Biophys.J., 110, 2016
|
|
3LGK
 
 | | D99N Epi-isozizaene synthase | | 分子名称: | Epi-isozizaene synthase, SULFATE ION | | 著者 | Aaron, J.A, Lin, X, Cane, D.E, Christianson, D.W. | | 登録日 | 2010-01-20 | | 公開日 | 2010-02-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.892 Å) | | 主引用文献 | Structure of Epi-Isozizaene Synthase from Streptomyces coelicolor A3(2), a Platform for New Terpenoid Cyclization Templates
Biochemistry, 49, 2010
|
|
3LHW
 
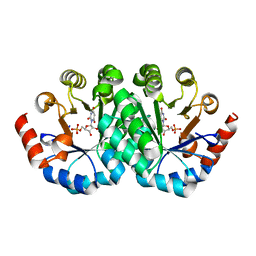 | | Crystal structure of the mutant V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-23 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LK2
 
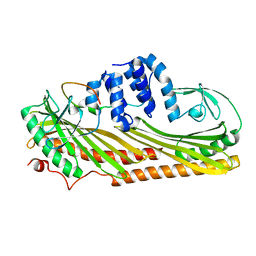 | | Crystal structure of CapZ bound to the uncapping motif from CARMIL | | 分子名称: | F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2, Leucine-rich repeat-containing protein 16A | | 著者 | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Aguda, A.H, Larsson, M, Cooper, J.A, Robinson, R.C. | | 登録日 | 2010-01-27 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3LTS
 
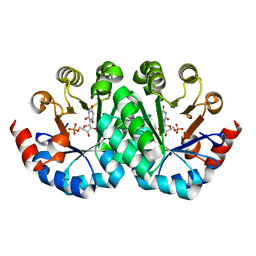 | | Crystal structure of the mutant V182A,I199A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-02-16 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.428 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3KRM
 
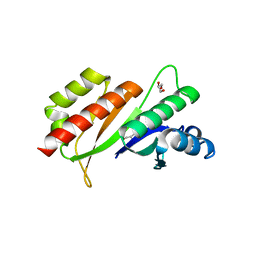 | | Imp1 kh34 | | 分子名称: | GLYCEROL, Insulin-like growth factor 2 mRNA-binding protein 1 | | 著者 | Chao, J.A, Singer, R.H, Almo, S.C, Patskovsky, Y. | | 登録日 | 2009-11-18 | | 公開日 | 2010-02-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | ZBP1 recognition of beta-actin zipcode induces RNA looping.
Genes Dev., 24, 2010
|
|
3KUM
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Arg-L-Tyr | | 分子名称: | ARGININE, Dipeptide Epimerase, MAGNESIUM ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | 登録日 | 2009-11-27 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5HEN
 
 | | Crystal structure of the N-terminus R100L bromodomain mutant of human BRD2 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2 | | 著者 | Tallant, C, Lori, C, Pasquo, A, Chiaraluce, R, Consalvi, V, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | 登録日 | 2016-01-06 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Crystal structure of the N-terminus R100L bromodomain mutant of human BRD2
To Be Published
|
|
5H9A
 
 | | Crystal structure of the Apo form of human cellular retinol binding protein 1 | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Retinol-binding protein 1 | | 著者 | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | 登録日 | 2015-12-26 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.381 Å) | | 主引用文献 | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
5HQJ
 
 | | Crystal structure of ABC transporter Solute Binding Protein B1G1H7 from Burkholderia graminis C4D1M, target EFI-511179, in complex with D-arabinose | | 分子名称: | CHLORIDE ION, Periplasmic binding protein/LacI transcriptional regulator, alpha-D-arabinopyranose | | 著者 | Roth, Y, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2016-01-21 | | 公開日 | 2016-03-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of ABC transporter Solute Binding Protein B1G1H7 from Burkholderia graminis C4D1M, target EFI-511179, in complex with D-arabinose
To be published
|
|
5HUR
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25T/I92K at cryogenic temperature | | 分子名称: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | 著者 | Skerritt, L.A, Caro, J.A, Heroux, A, Schlessman, J.L, Garcia-Moreno E, B. | | 登録日 | 2016-01-27 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25T/I92K at cryogenic temperature
To be Published
|
|
3L82
 
 | | X-ray Crystal structure of TRF1 and Fbx4 complex | | 分子名称: | F-box only protein 4, Telomeric repeat-binding factor 1 | | 著者 | Zeng, Z.X, Wang, W, Yang, Y.T, Chen, Y, Yang, X.M, Diehl, J.A, Liu, X.D, Lei, M. | | 登録日 | 2009-12-29 | | 公開日 | 2010-03-09 | | 最終更新日 | 2013-09-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis of Selective Ubiquitination of TRF1 by SCF(Fbx4)
Dev.Cell, 18, 2010
|
|
3LHV
 
 | | Crystal structure of the mutant V182A.I199A.V201A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-23 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LK3
 
 | | Crystal structure of CapZ bound to the CPI and CSI uncapping motifs from CARMIL | | 分子名称: | F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2, Leucine-rich repeat-containing protein 16A | | 著者 | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Cooper, J.A, Robinson, R.C. | | 登録日 | 2010-01-27 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3LHZ
 
 | | Crystal structure of the mutant V201A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-23 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LQE
 
 | |
3LHY
 
 | | Crystal structure of the mutant I199A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-23 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
