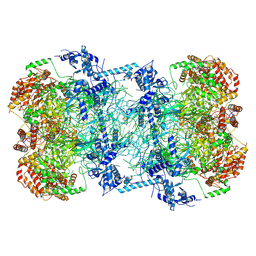
5UPW
 
 | |
4ES5
 
 | |
6U8Y
 
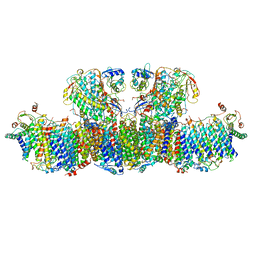 | |
8HIH
 
 | | Cryo-EM structure of Mycobacterium tuberculosis transcription initiation complex with transcription factor GlnR | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Lin, W, Shi, J, Xu, J.C. | | 登録日 | 2022-11-20 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Structural insights into the transcription activation mechanism of the global regulator GlnR from actinobacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2FA9
 
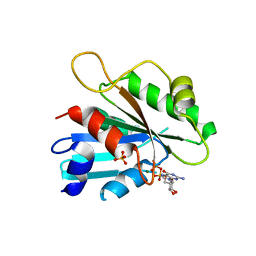 | | The crystal structure of Sar1[H79G]-GDP provides insight into the coat-controlled GTP hydrolysis in the disassembly of COP II | | 分子名称: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Rao, Y, Huang, M, Yuan, C, Bian, C, Hou, X. | | 登録日 | 2005-12-07 | | 公開日 | 2006-09-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of Sar1[H79G]-GDP Which Provides Insight into the Coat-controlled GTP Hydrolysis in the Disassembly of COP II
Chin.J.Struct.Chem., 25, 2006
|
|
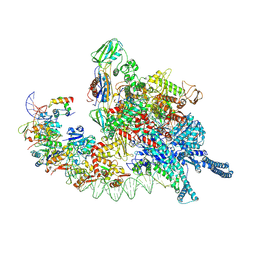
5BK4
 
 | | Cryo-EM structure of Mcm2-7 double hexamer on dsDNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (60-mer), strand 1, ... | | 著者 | Li, H, Yuan, Z, Bai, L. | | 登録日 | 2017-09-12 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3S6K
 
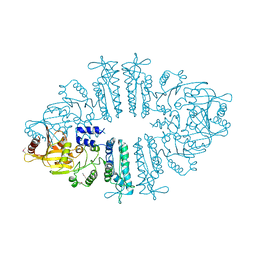 | | Crystal structure of xcNAGS | | 分子名称: | Acetylglutamate kinase | | 著者 | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | 登録日 | 2011-05-25 | | 公開日 | 2012-04-18 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.8018 Å) | | 主引用文献 | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3S6G
 
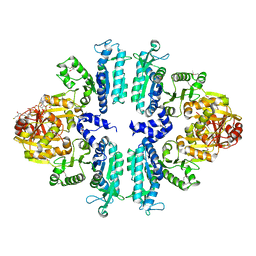 | | Crystal structures of Seleno-substituted mutant mmNAGS in space group P212121 | | 分子名称: | 1,2-ETHANEDIOL, COENZYME A, MALONATE ION, ... | | 著者 | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | 登録日 | 2011-05-25 | | 公開日 | 2012-04-18 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.6681 Å) | | 主引用文献 | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3S6H
 
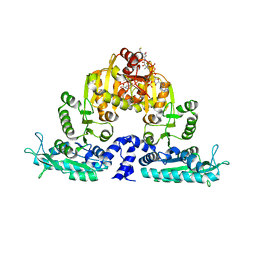 | | Crystal structure of native mmNAGS/k | | 分子名称: | COENZYME A, GLUTAMIC ACID, N-acetylglutamate kinase / N-acetylglutamate synthase | | 著者 | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | 登録日 | 2011-05-25 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
7ER3
 
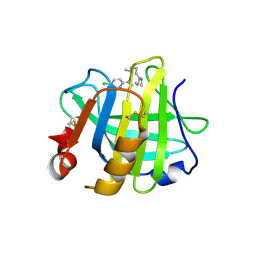 | | Crystal structure of beta-lactoglobulin complexed with chloroquine | | 分子名称: | (4S)-N~4~-(7-chloroquinolin-4-yl)-N~1~,N~1~-diethylpentane-1,4-diamine, Major allergen beta-lactoglobulin | | 著者 | Yao, Q, Ma, J, Xing, Y, Zang, J. | | 登録日 | 2021-05-05 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Binding of Chloroquine to Whey Protein Relieves Its Cytotoxicity while Enhancing Its Uptake by Cells.
J.Agric.Food Chem., 69, 2021
|
|
9MXF
 
 | | GRN-P4A isomer 1, granulin | | 分子名称: | Granulin peptide | | 著者 | Raffaelli, T, Daly, N. | | 登録日 | 2025-01-19 | | 公開日 | 2025-06-18 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Topological isomers of a potent wound healing peptide: structural insights and implications for bioactivity.
J.Biol.Chem., 2025
|
|
9MXE
 
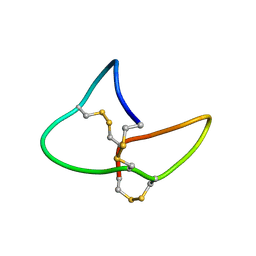 | | GRN-P4A isomer 2, granulin | | 分子名称: | Granulin peptide | | 著者 | Raffaelli, T, Daly, N. | | 登録日 | 2025-01-19 | | 公開日 | 2025-06-18 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Topological isomers of a potent wound healing peptide: structural insights and implications for bioactivity.
J.Biol.Chem., 2025
|
|
9MXG
 
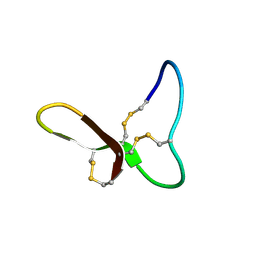 | | GRN-P4A mutant, granulin | | 分子名称: | Granulin peptide | | 著者 | Raffaelli, T, Daly, N. | | 登録日 | 2025-01-19 | | 公開日 | 2025-06-18 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Topological isomers of a potent wound healing peptide: structural insights and implications for bioactivity.
J.Biol.Chem., 2025
|
|
4WQ6
 
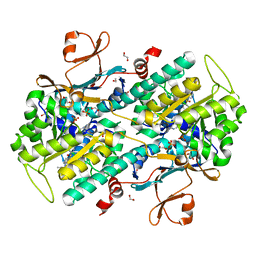 | | The crystal structure of human Nicotinamide phosphoribosyltransferase (NAMPT) in complex with N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide inhibitor (compound 21) | | 分子名称: | 1,2-ETHANEDIOL, N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Li, D, Wang, W. | | 登録日 | 2014-10-21 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Identification of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with no evidence of CYP3A4 time-dependent inhibition and improved aqueous solubility.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3QN7
 
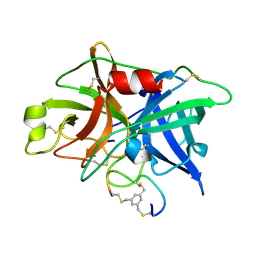 | | Potent and selective bicyclic peptide inhibitor (UK18) of human urokinase-type plasminogen activator(uPA) | | 分子名称: | 1,3,5-tris(bromomethyl)benzene, Bicyclic peptide inhibitor, Urokinase-type plasminogen activator | | 著者 | Angelini, A, Cendron, L, Touati, J, Winter, G, Zanotti, G, Heinis, C. | | 登録日 | 2011-02-08 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Bicyclic peptide inhibitor reveals large contact interface with a protease target
Acs Chem.Biol., 7, 2012
|
|
6S1K
 
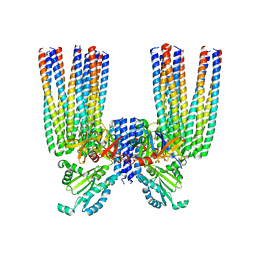 | | E. coli Core Signaling Unit, carrying QQQQ receptor mutation | | 分子名称: | CheW, Chemotaxis protein CheA, Methyl-accepting chemotaxis protein I | | 著者 | Cassidy, C.K. | | 登録日 | 2019-06-18 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (8.38 Å) | | 主引用文献 | Structure and dynamics of the E. coli chemotaxis core signaling complex by cryo-electron tomography and molecular simulations.
Commun Biol, 3, 2020
|
|
6PTJ
 
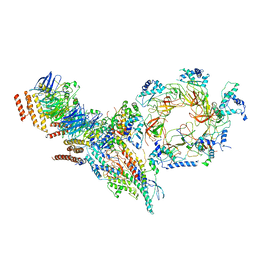 | | Structure of Ctf4 trimer in complex with one CMG helicase | | 分子名称: | Cell division control protein 45, DNA polymerase alpha-binding protein, DNA replication complex GINS protein PSF1, ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | 登録日 | 2019-07-15 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PTO
 
 | | Structure of Ctf4 trimer in complex with three CMG helicases | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | 登録日 | 2019-07-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PSY
 
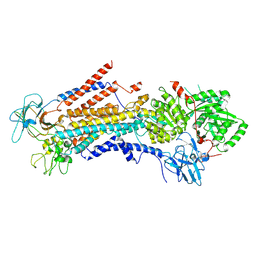 | | Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the autoinhibited apo form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | 著者 | Bai, L, Li, H. | | 登録日 | 2019-07-14 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p.
Nat Commun, 10, 2019
|
|
6PTN
 
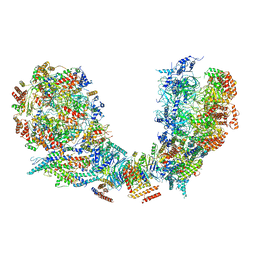 | | Structure of Ctf4 trimer in complex with two CMG helicases | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | 登録日 | 2019-07-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PSX
 
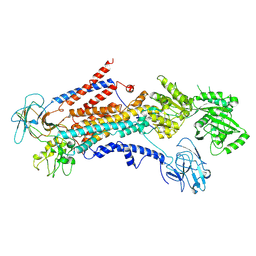 | | Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the PI4P-activated form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | 著者 | Bai, L, Li, H. | | 登録日 | 2019-07-14 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p.
Nat Commun, 10, 2019
|
|
6YJ5
 
 | |
3BLI
 
 | |
3BLE
 
 | |
5KIT
 
 | |
