8V74
 
 | |
8V6Y
 
 | |
8V76
 
 | |
8V78
 
 | |
5VJ6
 
 | |
8H1M
 
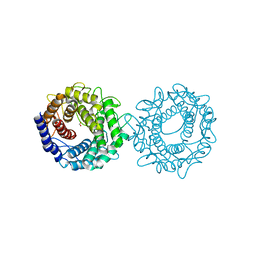 | | Crystal structure of glucose-2-epimerase mutant_D254A from Runella slithyformis Runsl_4512 | | 分子名称: | FORMIC ACID, N-acylglucosamine 2-epimerase | | 著者 | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | 登録日 | 2022-10-03 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1N
 
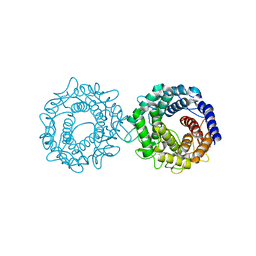 | | Crystal structure of glucose-2-epimerase mutant_D254A in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | 分子名称: | FORMIC ACID, N-acylglucosamine 2-epimerase, sorbitol | | 著者 | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | 登録日 | 2022-10-03 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5W2I
 
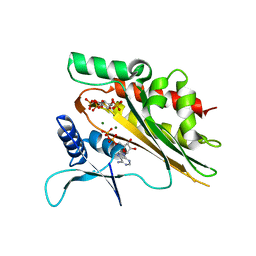 | | Crystal structure of the core catalytic domain of human inositol phosphate multikinase soaked with C4-analogue of PtdIns(4,5)P2 and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol polyphosphate multikinase,Inositol polyphosphate multikinase, ... | | 著者 | Wang, H, Shears, S.B. | | 登録日 | 2017-06-06 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural features of human inositol phosphate multikinase rationalize its inositol phosphate kinase and phosphoinositide 3-kinase activities.
J. Biol. Chem., 292, 2017
|
|
5W2H
 
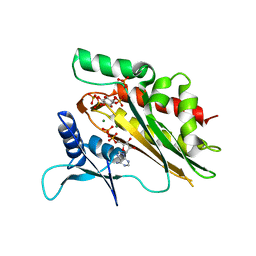 | | Crystal structure of the core catalytic domain of human inositol phosphate multikinase in complex with Ins(1,4,5)P3 and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol polyphosphate multikinase,Inositol polyphosphate multikinase, ... | | 著者 | Wang, H, Shears, S.B. | | 登録日 | 2017-06-06 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural features of human inositol phosphate multikinase rationalize its inositol phosphate kinase and phosphoinositide 3-kinase activities.
J. Biol. Chem., 292, 2017
|
|
7ZSA
 
 | |
7ZSB
 
 | |
7ZS9
 
 | |
3T9A
 
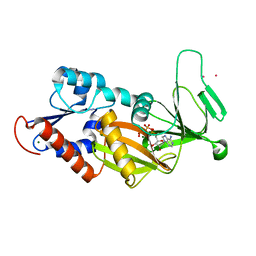 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP at pH 7.0 | | 分子名称: | CADMIUM ION, Inositol Pyrophosphate Kinase, MAGNESIUM ION, ... | | 著者 | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | 登録日 | 2011-08-02 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T9E
 
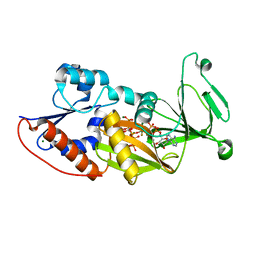 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP, 5-(PP)-IP5 (5-IP7) and MgF3 (transition state mimic) | | 分子名称: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-DIPHOSPHATE, Inositol Pyrophosphate Kinase, ... | | 著者 | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | 登録日 | 2011-08-02 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T54
 
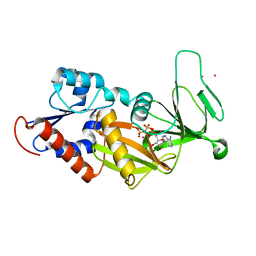 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ATP and Cadmium | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CADMIUM ION, Inositol Pyrophosphate Kinase | | 著者 | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | 登録日 | 2011-07-26 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T9B
 
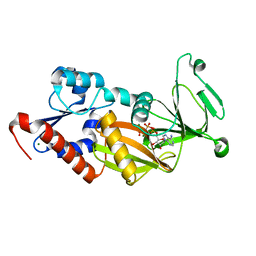 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP at pH 5.2 | | 分子名称: | Inositol Pyrophosphate Kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | 登録日 | 2011-08-02 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T9D
 
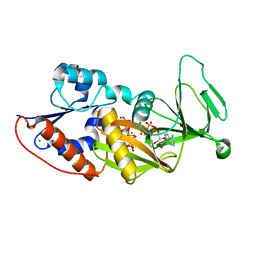 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP and 5-(PP)-IP5 (5-IP7) | | 分子名称: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Inositol Pyrophosphate Kinase, MAGNESIUM ION, ... | | 著者 | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | 登録日 | 2011-08-02 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
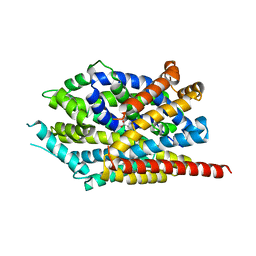
3USI
 
 | |
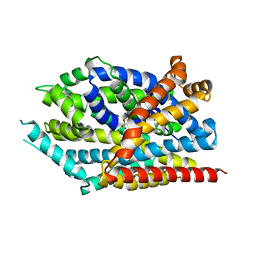
3USK
 
 | |
3USL
 
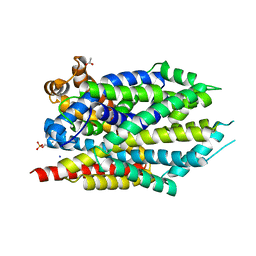 | | Crystal Structure of LeuT bound to L-selenomethionine in space group C2 from lipid bicelles | | 分子名称: | ACETATE ION, IODIDE ION, PHOSPHOCHOLINE, ... | | 著者 | Wang, H, Elferich, J, Gouaux, E. | | 登録日 | 2011-11-23 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3T7A
 
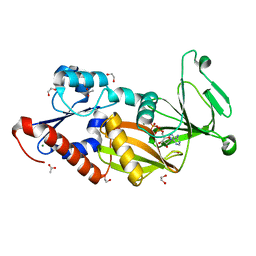 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP at pH 5.2 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | 登録日 | 2011-07-29 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T9F
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and 1,5-(PP)2-IP4 (1,5-IP8) | | 分子名称: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], ADENOSINE-5'-DIPHOSPHATE, CADMIUM ION, ... | | 著者 | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | 登録日 | 2011-08-02 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T9C
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP and inositol hexakisphosphate (IP6) | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, Inositol Pyrophosphate Kinase, MAGNESIUM ION, ... | | 著者 | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | 登録日 | 2011-08-02 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
5YXA
 
 | | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus | | 分子名称: | Non-structural protein 1 | | 著者 | Wang, H, Song, H, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2017-12-04 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus.
Sci China Life Sci, 60, 2017
|
|
3USO
 
 | |
