3HTE
 
 | | Crystal structure of nucleotide-free hexameric ClpX | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit clpX, SULFATE ION | | 著者 | Glynn, S.E, Martin, A, Baker, T.A, Sauer, R.T. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (4.026 Å) | | 主引用文献 | Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine.
Cell(Cambridge,Mass.), 139, 2009
|
|
3HWS
 
 | | Crystal structure of nucleotide-bound hexameric ClpX | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit clpX, MAGNESIUM ION, ... | | 著者 | Glynn, S.E, Martin, A, Baker, T.A, Sauer, R.T. | | 登録日 | 2009-06-18 | | 公開日 | 2009-11-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine.
Cell(Cambridge,Mass.), 139, 2009
|
|
6J06
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with HMBPP-08 | | 分子名称: | (2E)-3-(hydroxymethyl)-4-(4-methylphenyl)but-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A1, CALCIUM ION, ... | | 著者 | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | 登録日 | 2018-12-21 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
3LGY
 
 | |
6J03
 
 | | Crystal structure of a cyclase mutant in apo form | | 分子名称: | 12-epi-hapalindole U synthase, CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium | | 著者 | Tang, X.K, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2018-12-20 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Structural insights into the calcium dependence of Stig cyclases.
Rsc Adv, 9, 2019
|
|
3LQX
 
 | |
3IPN
 
 | | Crystal Structure of fluorine and methyl modified collagen: (mepFlpgly)7 | | 分子名称: | CARBONATE ION, Non-natural Collagen | | 著者 | Satyshur, K.A, Shoulders, M.D, Raines, R.T, Forest, K.T. | | 登録日 | 2009-08-18 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Stereoelectronic and steric effects in side chains preorganize a protein main chain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6J8V
 
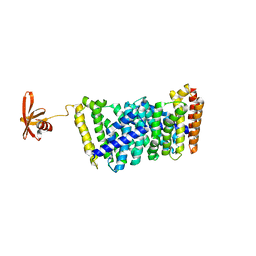 | | Structure of MOEN5-SSO7D fusion protein in complex with ligand 2 | | 分子名称: | FARNESYL, MoeN5,DNA-binding protein 7d | | 著者 | Ko, T.P, Zhang, L.L, Chen, C.C, Guo, R.T. | | 登録日 | 2019-01-21 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
3LGV
 
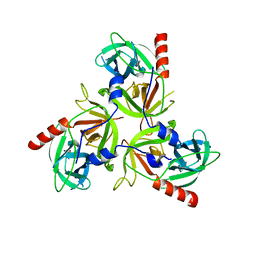 | |
3LGW
 
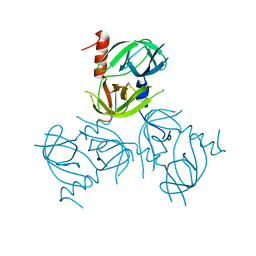 | |
3LGI
 
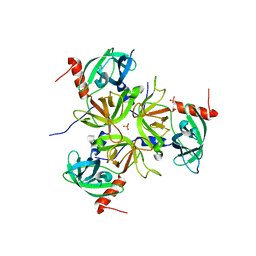 | |
3LGT
 
 | |
3LH3
 
 | | DFP modified DegS delta PDZ | | 分子名称: | Protease degS | | 著者 | Sohn, J, Grant, R.A, Sauer, R.T. | | 登録日 | 2010-01-21 | | 公開日 | 2010-08-25 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
4HW0
 
 | | Crystal structure of Sso10a-2, a DNA-binding protein from Sulfolobus solfataricus | | 分子名称: | DNA-binding protein Sso10a-2 | | 著者 | Waterreus, W.J, Goosen, N, Moolenaar, G.F, Driessen, R.P.C, Dame, R.T, Pannu, N.S. | | 登録日 | 2012-11-07 | | 公開日 | 2013-10-30 | | 最終更新日 | 2017-01-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Diverse architectural properties of Sso10a proteins: Evidence for a role in chromatin compaction and organization.
Sci Rep, 6, 2016
|
|
4I63
 
 | | Crystal Structure of E-R ClpX Hexamer | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | 著者 | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | 登録日 | 2012-11-29 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (5.709 Å) | | 主引用文献 | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
3RSD
 
 | |
4FRN
 
 | | Crystal structure of the cobalamin riboswitch regulatory element | | 分子名称: | BARIUM ION, Cobalamin riboswitch aptamer domain, Hydroxocobalamin | | 著者 | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | 登録日 | 2012-06-26 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.43 Å) | | 主引用文献 | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
4FRG
 
 | | Crystal structure of the cobalamin riboswitch aptamer domain | | 分子名称: | Hydroxocobalamin, IRIDIUM (III) ION, MAGNESIUM ION, ... | | 著者 | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | 登録日 | 2012-06-26 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
4GMA
 
 | | Crystal structure of the adenosylcobalamin riboswitch | | 分子名称: | Adenosylcobalamin, Adenosylcobalamin riboswitch | | 著者 | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | 登録日 | 2012-08-15 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.94 Å) | | 主引用文献 | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
4ISZ
 
 | | RNA ligase RtcB in complex with GTP alphaS and Mn(II) | | 分子名称: | GUANOSINE-5'-RP-ALPHA-THIO-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | 著者 | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | 登録日 | 2013-01-17 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
4IWN
 
 | | Crystal structure of a putative methyltransferase CmoA in complex with a novel SAM derivative | | 分子名称: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, tRNA (cmo5U34)-methyltransferase | | 著者 | Aller, P, Lobley, C.M, Byrne, R.T, Antson, A.A, Waterman, D.G. | | 登録日 | 2013-01-24 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | S-Adenosyl-S-carboxymethyl-L-homocysteine: a novel cofactor found in the putative tRNA-modifying enzyme CmoA.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7URT
 
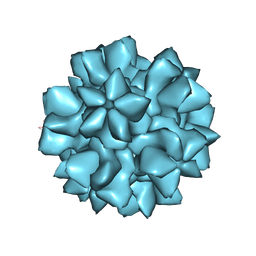 | |
7URN
 
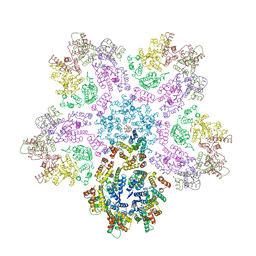 | |
4G34
 
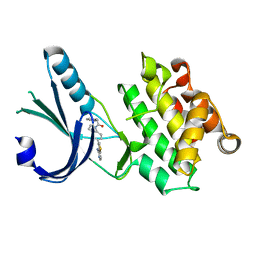 | | Crystal Structure of GSK6924 Bound to PERK (R587-R1092, delete A660-T867) at 2.70 A Resolution | | 分子名称: | 1-[5-(4-aminothieno[3,2-c]pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]-2-phenylethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | 著者 | Gampe, R.T, Axten, J.M. | | 登録日 | 2012-07-13 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
4G31
 
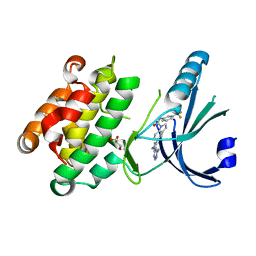 | | Crystal Structure of GSK6414 Bound to PERK (R587-R1092, delete A660-T867) at 2.28 A Resolution | | 分子名称: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-2,3-dihydro-1H-indol-1-yl]-2-[3-(trifluoromethyl)phenyl]ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL | | 著者 | Gampe, R.T, Axten, J.M. | | 登録日 | 2012-07-13 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
