8OOK
 
 | |
8OO9
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-DNA refinement state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase INO80, DNA strand 1, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOT
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state2 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOA
 
 | | CryoEM Structure INO80core Hexasome complex Hexasome refinement state1 | | 分子名称: | DNA Strand 2, DNA strand 1, Histone H2A, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOP
 
 | | CryoEM Structure INO80core Hexasome complex composite model state2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOF
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state1 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OO7
 
 | | CryoEM Structure INO80core Hexasome complex composite model state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OVR
 
 | | Clostridium perfringens chitinase CP56_3454 apo form | | 分子名称: | Chitinase B, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), SODIUM ION, ... | | 著者 | Bloch, Y, Savvides, S.N. | | 登録日 | 2023-04-26 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Clostridium perfringens chitinases, key enzymes during early stages of necrotic enteritis in broiler chickens.
Plos Pathog., 20, 2024
|
|
4TNJ
 
 | | RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 4.5 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | 登録日 | 2014-06-04 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNI
 
 | | RT XFEL structure of Photosystem II 500 ms after the third illumination at 4.6 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | 登録日 | 2014-06-04 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (4.6 Å) | | 主引用文献 | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNK
 
 | | RT XFEL structure of Photosystem II 250 microsec after the third illumination at 5.2 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | 登録日 | 2014-06-04 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (5.2 Å) | | 主引用文献 | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
2X0N
 
 | | Structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei determined from Laue data | | 分子名称: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, GLYCOSOMAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Vellieux, F.M.D, Hajdu, J, Hol, W.G.J. | | 登録日 | 2009-12-16 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma Brucei Determined from Laue Data.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
4TNH
 
 | | RT XFEL structure of Photosystem II in the dark state at 4.9 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | 登録日 | 2014-06-04 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (4.900007 Å) | | 主引用文献 | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNL
 
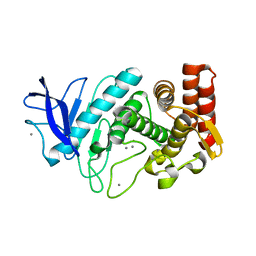 | | 1.8 A resolution room temperature structure of Thermolysin recorded using an XFEL | | 分子名称: | CALCIUM ION, Thermolysin, ZINC ION | | 著者 | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | 登録日 | 2014-06-04 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
7SSF
 
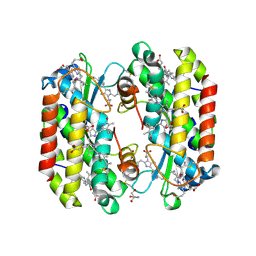 | | Light harvesting phycobiliprotein HaPE560 from the cryptophyte Hemiselmis andersenii CCMP644 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DiCys-(15,16)-Dihydrobiliverdin, ... | | 著者 | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | 登録日 | 2021-11-10 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii
Commun Biol, 2023
|
|
7SUT
 
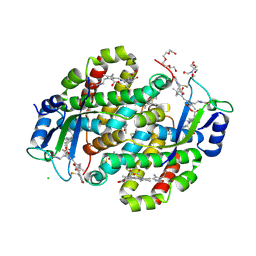 | | Light harvesting phycobiliprotein HaPE645 from the cryptophyte Hemiselmis andersenii CCMP644 | | 分子名称: | (15,16)-DIHYDROBILIVERDIN (SINGLY LINKED), 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | 登録日 | 2021-11-18 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
7Z3T
 
 | | Crystal structure of apo human Cathepsin L | | 分子名称: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | 登録日 | 2022-03-02 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7YOO
 
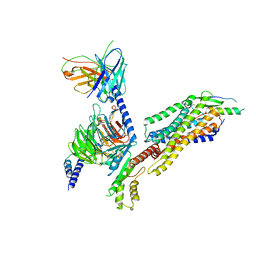 | | Complex structure of Neuropeptide Y Y2 receptor in complex with NPY and Gi | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Kang, H, Park, C, Kim, J, Choi, H.-J. | | 登録日 | 2022-08-01 | | 公開日 | 2023-03-22 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structural basis for Y2 receptor-mediated neuropeptide Y and peptide YY signaling.
Structure, 31, 2023
|
|
7Z58
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound Calpeptin | | 分子名称: | 1,2-ETHANEDIOL, Calpeptin, Cathepsin L, ... | | 著者 | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | 登録日 | 2022-03-08 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7ZAK
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | 登録日 | 2022-03-22 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
7VYO
 
 | | The structure of GdmN | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | 登録日 | 2021-11-14 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7ZGE
 
 | |
5HLL
 
 | |
7LM4
 
 | | The crystal structure of the I38T mutant PA Endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | 分子名称: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | 著者 | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | 登録日 | 2021-02-05 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
