4OQ3
 
 | |
5CCP
 
 | | HISTIDINE 52 IS A CRITICAL RESIDUE FOR RAPID FORMATION OF CYTOCHROME C PEROXIDASE COMPOUND I | | 分子名称: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Brown, K, Shaw, A, Miller, M.A, Kraut, J. | | 登録日 | 1993-06-07 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Histidine 52 is a critical residue for rapid formation of cytochrome c peroxidase compound I.
Biochemistry, 32, 1993
|
|
6C1S
 
 | | Phosphoinositide 3-Kinase gamma bound to an pyrrolopyridinone Inhibitor | | 分子名称: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, {4-[2-(5,6-dimethoxypyridin-3-yl)-5-oxo-5,7-dihydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-1H-pyrazol-1-yl}acetonitrile | | 著者 | Jacobs, M.D, Griffin, J.P. | | 登録日 | 2018-01-05 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Design and Synthesis of a Novel Series of Orally Bioavailable, CNS-Penetrant, Isoform Selective Phosphoinositide 3-Kinase gamma (PI3K gamma ) Inhibitors with Potential for the Treatment of Multiple Sclerosis (MS).
J. Med. Chem., 61, 2018
|
|
3EKO
 
 | |
3EE1
 
 | |
1JQM
 
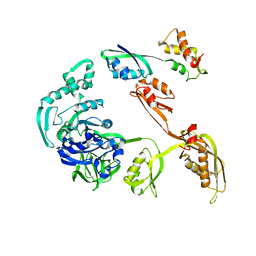 | | Fitting of L11 protein and elongation factor G (EF-G) in the cryo-em map of e. coli 70S ribosome bound with EF-G, GDP and fusidic acid | | 分子名称: | 50S Ribosomal protein L11, Elongation Factor G | | 著者 | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | 登録日 | 2001-08-07 | | 公開日 | 2001-09-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
6WIO
 
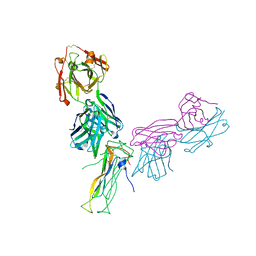 | | Fab antigen complex | | 分子名称: | Fab Heavy chain, Fab Light chain, Interleukin-17A | | 著者 | Antonysamy, S. | | 登録日 | 2020-04-10 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Rapid and robust antibody Fab fragment crystallization utilizing edge-to-edge beta-sheet packing.
Plos One, 15, 2020
|
|
6A06
 
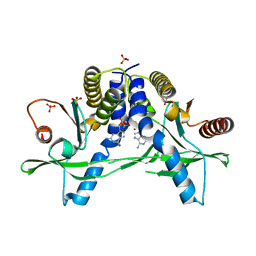 | | Structure of pSTING complex | | 分子名称: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A04
 
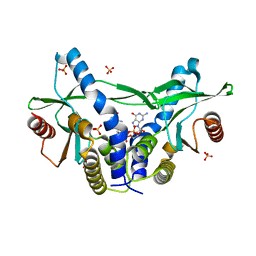 | | Structure of pSTING complex | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6XGR
 
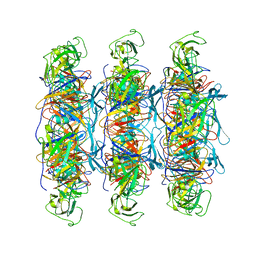 | | YSD1 major tail protein | | 分子名称: | YSD1_22 major tail protein | | 著者 | Hardy, J.M, Dunstan, R, Venugopal, H, Lithgow, T.J, Coulibaly, F.J. | | 登録日 | 2020-06-17 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The architecture and stabilisation of flagellotropic tailed bacteriophages.
Nat Commun, 11, 2020
|
|
8K6L
 
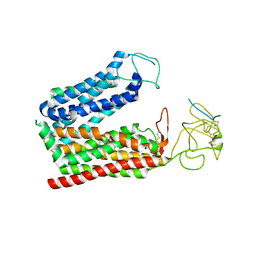 | | Cryo-EM structure of human OATP1B1 in complex with DCF | | 分子名称: | 2',7'-bis(chloranyl)-3',6'-bis(oxidanyl)spiro[2-benzofuran-3,9'-xanthene]-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Shan, Z, Yang, X, Zhang, Y. | | 登録日 | 2023-07-25 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
6XGP
 
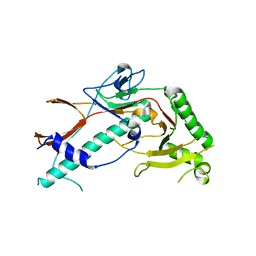 | | YSD1_17 major capsid protein | | 分子名称: | YSD1_17 major capsid protein | | 著者 | Grinter, R, Hardy, J.M, Dunstan, R, Lithgow, T.J, Coulibaly, F.J. | | 登録日 | 2020-06-17 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The architecture and stabilisation of flagellotropic tailed bacteriophages.
Nat Commun, 11, 2020
|
|
8G8C
 
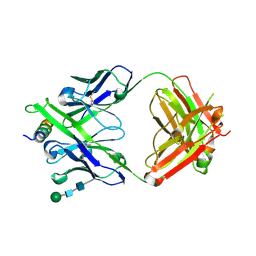 | | Crystal structure of DH1322.1 Fab in complex with HIV proximal MPER peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1322.1 heavy chain, DH1322.1 light chain, ... | | 著者 | Niyongabo, A, Janus, B.M, Ofek, G. | | 登録日 | 2023-02-17 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
6A05
 
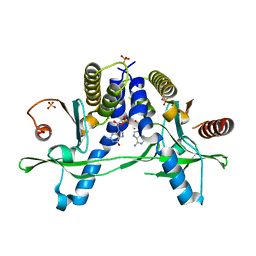 | | Structure of pSTING complex | | 分子名称: | 2-amino-9-[(2R,3R,3aR,5S,7aS,9R,10R,10aR,12R,14aS)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A03
 
 | | Structure of pSTING complex | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.597 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6KZ4
 
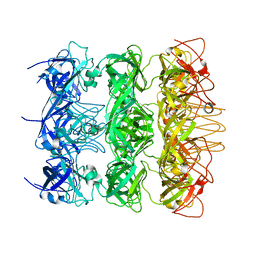 | | YebT domain 5-7 | | 分子名称: | Intermembrane transport protein YebT | | 著者 | Wang, H.W, Liu, C, Zhang, L. | | 登録日 | 2019-09-23 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM Structure of a Bacterial Lipid Transporter YebT.
J.Mol.Biol., 432, 2020
|
|
6KYB
 
 | | Crystal structure of Atg18 from Saccharomyces cerevisiae | | 分子名称: | Autophagy-related protein 18 | | 著者 | Tang, D, Lei, Y, Liao, G, Chen, Q, Xu, L, Lu, K, Qi, S. | | 登録日 | 2019-09-17 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of Atg18 reveals a new binding site for Atg2 in Saccharomyces cerevisiae.
Cell.Mol.Life Sci., 78, 2021
|
|
6KZ3
 
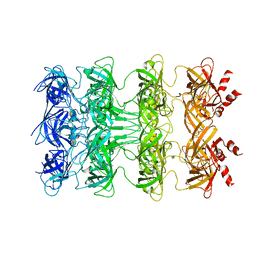 | | YebT domain1-4 | | 分子名称: | Intermembrane transport protein YebT | | 著者 | Wang, H.W, Liu, C, Zhang, L. | | 登録日 | 2019-09-23 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM Structure of a Bacterial Lipid Transporter YebT.
J.Mol.Biol., 432, 2020
|
|
6ENE
 
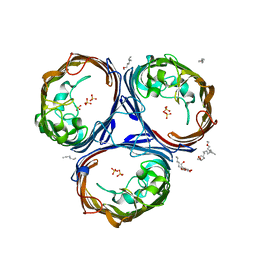 | | OmpF orthologue from Enterobacter cloacae (OmpE35) | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein (Porin), SULFATE ION, ... | | 著者 | van den Berg, B, Abellon-Ruiz, J, Basle, A. | | 登録日 | 2017-10-04 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
7DRS
 
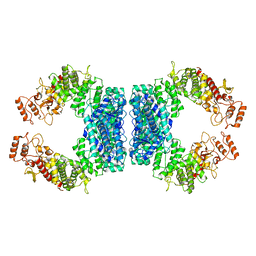 | | Structure of SspE_40224 | | 分子名称: | SspE protein | | 著者 | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | 登録日 | 2020-12-29 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.42 Å) | | 主引用文献 | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7DRR
 
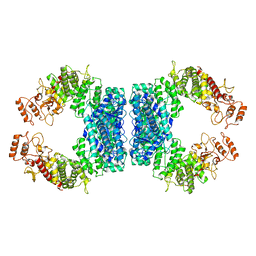 | | Structure of SspE-R100A protein | | 分子名称: | SspE protein | | 著者 | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | 登録日 | 2020-12-29 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.48 Å) | | 主引用文献 | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7E7Y
 
 | |
7E7X
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E88
 
 | |
