7VPD
 
 | |
7VO0
 
 | |
7VO9
 
 | |
7VPZ
 
 | |
7XGL
 
 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in Apo form | | 分子名称: | CHLORIDE ION, GLYCEROL, Quinolinate Phosphoribosyl Transferase, ... | | 著者 | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | 登録日 | 2022-04-05 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7XGN
 
 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in complex with Nicotinic Acid (NA) | | 分子名称: | CHLORIDE ION, NICOTINIC ACID, Quinolinate Phosphoribosyl Transferase, ... | | 著者 | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | 登録日 | 2022-04-05 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7XGM
 
 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in complex with Quinolinic Acid (QA) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, QUINOLINIC ACID, ... | | 著者 | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | 登録日 | 2022-04-05 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8H89
 
 | | Capsid of Ralstonia phage GP4 | | 分子名称: | Major capsid protein, Virion associated protein | | 著者 | Liu, H.R, Chen, W.Y. | | 登録日 | 2022-10-22 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | A Capsid Structure of Ralstonia solanacearum podoviridae GP4 with a Triangulation Number T = 9.
Viruses, 14, 2022
|
|
8XAR
 
 | | Structure-Based Design and Optimization of Methionine Adenosyltransferase 2A (MAT2A) Inhibitors with SAM and Compound 54 | | 分子名称: | 1,2-ETHANEDIOL, 7-chloranyl-2-ethyl-5-pyridin-3-yl-pyrazolo[3,4-c]quinolin-4-one, CHLORIDE ION, ... | | 著者 | Zheng, J.Y, Zhang, G.P, Li, J.J, Tong, S.L. | | 登録日 | 2023-12-05 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | Structure-Based Design and Optimization of Methionine Adenosyltransferase 2A (MAT2A) Inhibitors with High Selectivity, Brain Penetration, and In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
8XB0
 
 | | Structure-Based Design and Optimization of Methionine Adenosyltransferase 2A (MAT2A) Inhibitors with SAM and Compound 292 | | 分子名称: | 1,2-ETHANEDIOL, 7-chloranyl-5-(2-cyclopropylpyridin-3-yl)-8-fluoranyl-2-methyl-pyrazolo[3,4-c]quinolin-4-one, CHLORIDE ION, ... | | 著者 | Tong, S.L, Zhang, G.P. | | 登録日 | 2023-12-05 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Structure-Based Design and Optimization of Methionine Adenosyltransferase 2A (MAT2A) Inhibitors with High Selectivity, Brain Penetration, and In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
6NKO
 
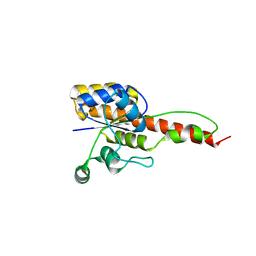 | | Crystal structure of ForH | | 分子名称: | ForH | | 著者 | Zheng, J, Irani, S, Zhang, Y. | | 登録日 | 2019-01-07 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.403 Å) | | 主引用文献 | Identification of the Formycin A Biosynthetic Gene Cluster from Streptomyces kaniharaensis Illustrates the Interplay between Biological Pyrazolopyrimidine Formation and de Novo Purine Biosynthesis.
J. Am. Chem. Soc., 141, 2019
|
|
8JOV
 
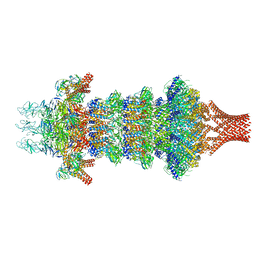 | | Portal-tail complex of phage GP4 | | 分子名称: | Portal protein, Putative tail fiber protein, Virion associated protein, ... | | 著者 | Liu, H, Chen, W. | | 登録日 | 2023-06-08 | | 公開日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Asymmetric Structure of Podophage GP4 Reveals a Novel Architecture of Three Types of Tail Fibers.
J.Mol.Biol., 435, 2023
|
|
8JOU
 
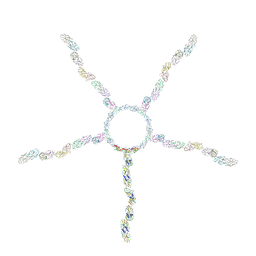 | | Fiber I and fiber-tail-adaptor of phage GP4 | | 分子名称: | Virion-associated phage protein, rope protein of phage GP4 | | 著者 | Liu, H, Chen, W. | | 登録日 | 2023-06-08 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Asymmetric Structure of Podophage GP4 Reveals a Novel Architecture of Three Types of Tail Fibers.
J.Mol.Biol., 435, 2023
|
|
3DVD
 
 | |
3DVB
 
 | |
3DVC
 
 | |
3DV7
 
 | |
5V39
 
 | |
7SLQ
 
 | | Cryo-EM structure of 7SK core RNP with circular RNA | | 分子名称: | 7SK snRNA methylphosphate capping enzyme, La-related protein 7, Minimal circular 7SK RNA, ... | | 著者 | Yang, Y, Liu, S, Zhou, Z.H, Feigon, J. | | 登録日 | 2021-10-24 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.
Mol.Cell, 82, 2022
|
|
7SLP
 
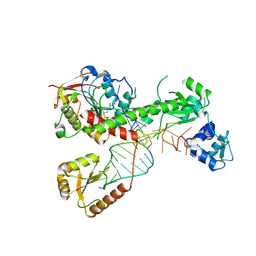 | | Cryo-EM structure of 7SK core RNP with linear RNA | | 分子名称: | 7SK snRNA methylphosphate capping enzyme, La-related protein 7, Linear 7SK RNA, ... | | 著者 | Yang, Y, Liu, S, Zhou, Z.H, Feigon, J. | | 登録日 | 2021-10-24 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.
Mol.Cell, 82, 2022
|
|
2QJU
 
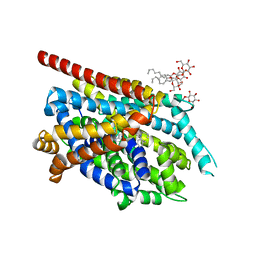 | | Crystal Structure of an NSS Homolog with Bound Antidepressant | | 分子名称: | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, CHLORIDE ION, LEUCINE, ... | | 著者 | Zhou, Z, Karpowich, N.K, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2007-07-09 | | 公開日 | 2007-08-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake.
Science, 317, 2007
|
|
5C3U
 
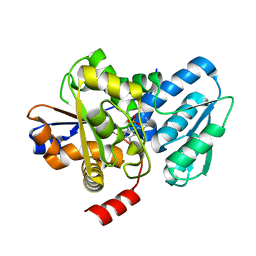 | | Crystal structure of a fungal L-serine ammonia-lyase from Rhizomucor miehei | | 分子名称: | L-serine ammonia-lyase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Zhen, Q, Qiaojuan, Y, Shaoqing, Y, Zhengqiang, J. | | 登録日 | 2015-06-17 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure and characterization of a novel l-serine ammonia-lyase from Rhizomucor miehei.
Biochem.Biophys.Res.Commun., 466, 2015
|
|
7MBO
 
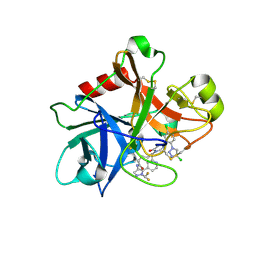 | | FACTOR XIA (PICHIA PASTORIS; C500S [C122S]) IN COMPLEX WITH THE INHIBITOR Milvexian (BMS-986177), IUPAC NAME:(6R,10S)-10-{4-[5-chloro-2-(4-chloro-1H-1,2,3-triazol-1-yl)phenyl]-6- oxopyrimidin-1(6H)-yl}-1-(difluoromethyl)-6-methyl-1,4,7,8,9,10-hexahydro-15,11- (metheno)pyrazolo[4,3-b][1,7]diazacyclotetradecin-5(6H)-one | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, Milvexian | | 著者 | Sheriff, S. | | 登録日 | 2021-04-01 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.924 Å) | | 主引用文献 | Discovery of Milvexian, a High-Affinity, Orally Bioavailable Inhibitor of Factor XIa in Clinical Studies for Antithrombotic Therapy.
J.Med.Chem., 65, 2022
|
|
6W50
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR METHYL ((10R,14S)- 14-(4-(3-CHLORO-2,6-DIFLUOROPHENYL)-6-OXO-3,6-DIHYDRO- 1(2H)-PYRIDINYL)-10-METHYL-9-OXO-8,16- DIAZATRICYCLO[13.3.1.0~2,7~]NONADECA-1(19),2,4,6,15,17- HEXAEN-5-YL)CARBAMATE | | 分子名称: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | 著者 | Sheriff, S. | | 登録日 | 2020-03-12 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery of a High Affinity, Orally Bioavailable Macrocyclic FXIa Inhibitor with Antithrombotic Activity in Preclinical Species.
J.Med.Chem., 63, 2020
|
|
6DHA
 
 | |
