6R83
 
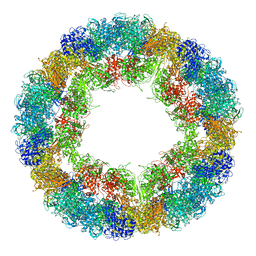 | | CryoEM structure and molecular model of squid hemocyanin (Todarodes pacificus , TpH) | | 分子名称: | Hemocyanin subunit 1 | | 著者 | Tanaka, Y, Kato, S, Stabrin, M, Raunser, S, Matsui, T, Gatsogiannis, C. | | 登録日 | 2019-03-31 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Cryo-EM reveals the asymmetric assembly of squid hemocyanin.
Iucrj, 6, 2019
|
|
1WTM
 
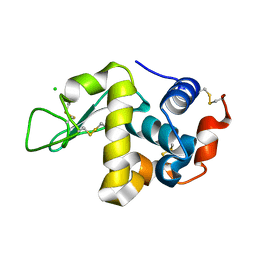 | | X-ray structure of HEW Lysozyme Orthorhombic Crystal formed in the Earth's magnetic field | | 分子名称: | CHLORIDE ION, Lysozyme C | | 著者 | Saijo, S, Yamada, Y, Sato, T, Tanaka, N, Matsui, T, Sazaki, G, Nakajima, K, Matsuura, Y. | | 登録日 | 2004-11-25 | | 公開日 | 2004-12-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Structural consequences of hen egg-white lysozyme orthorhombic crystal growth in a high magnetic field: validation of X-ray diffraction intensity, conformational energy searching and quantitative analysis of B factors and mosaicity.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5B0D
 
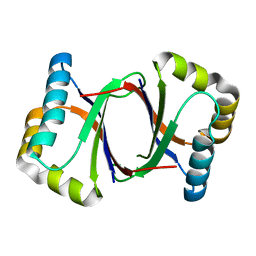 | | Polyketide cyclase OAC from Cannabis sativa, Y27W mutant | | 分子名称: | Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0C
 
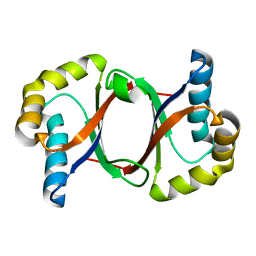 | | Polyketide cyclase OAC from Cannabis sativa, Y27F mutant | | 分子名称: | Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0B
 
 | | Polyketide cyclase OAC from Cannabis sativa, I7F mutant | | 分子名称: | ACETATE ION, Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.187 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
1UG7
 
 | | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806 | | 分子名称: | 2610208M17Rik protein | | 著者 | Li, H, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Shirouzu, M, Terada, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-06-13 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806
To be Published
|
|
5B09
 
 | | Polyketide cyclase OAC from Cannabis sativa bound with Olivetolic acid | | 分子名称: | 2,4-bis(oxidanyl)-6-pentyl-benzoic acid, Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0G
 
 | | Polyketide cyclase OAC from Cannabis sativa, H78S mutant | | 分子名称: | Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B08
 
 | | Polyketide cyclase OAC from Cannabis sativa | | 分子名称: | Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.325 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0A
 
 | | Polyketide cyclase OAC from Cannabis sativa, H5Q mutant | | 分子名称: | Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
1UH6
 
 | | Solution Structure of the murine ubiquitin-like 5 protein from RIKEN cDNA 0610031K06 | | 分子名称: | ubiquitin-like 5 | | 著者 | Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-06-25 | | 公開日 | 2003-12-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the murine ubiquitin-like 5 protein from RIKEN cDNA 0610031K06
To be Published
|
|
1UHS
 
 | | Solution structure of mouse homeodomain-only protein HOP | | 分子名称: | homeodomain only protein | | 著者 | Saito, K, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-07-10 | | 公開日 | 2004-07-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of mouse homeodomain-only protein HOP
To be Published
|
|
5B0E
 
 | | Polyketide cyclase OAC from Cannabis sativa, V59M mutant | | 分子名称: | GLYCEROL, Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.603 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
1UHR
 
 | | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a | | 分子名称: | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily D member 1 | | 著者 | Yamada, K, Saito, K, Nameki, N, Inoue, M, Koshiba, S, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-07-09 | | 公開日 | 2004-08-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a
To be Published
|
|
1UHT
 
 | | Solution Structure of The FHA Domain of Arabidopsis thaliana Hypothetical Protein | | 分子名称: | expressed protein | | 著者 | Tomizawa, T, Inoue, M, Koshiba, S, Hayashi, F, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Shinozaki, K, Seki, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-07-10 | | 公開日 | 2004-01-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of The FHA Domain of Arabidopsis thaliana Hypothetical Protein
To be Published
|
|
1UG2
 
 | | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain | | 分子名称: | 2610100B20Rik gene product | | 著者 | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-06-11 | | 公開日 | 2004-06-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain
To be Published
|
|
6JUX
 
 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with RK-71807 | | 分子名称: | 4-(1-ethyl-3-pyridin-3-yl-pyrazol-4-yl)-~{N}-(4-piperazin-1-ylphenyl)pyrimidin-2-amine, Activin receptor type-1, SULFATE ION | | 著者 | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | 登録日 | 2019-04-15 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis of Activin Receptor-Like Kinase 2 (R206H) Inhibition by Bis-heteroaryl Pyrazole-Based Inhibitors for the Treatment of Fibrodysplasia Ossificans Progressiva Identified by the Integration of Ligand-Based and Structure-Based Drug Design Approaches.
Acs Omega, 5, 2020
|
|
5B0F
 
 | | Polyketide cyclase OAC from Cannabis sativa, Y72F mutant | | 分子名称: | GLYCEROL, Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
1DIO
 
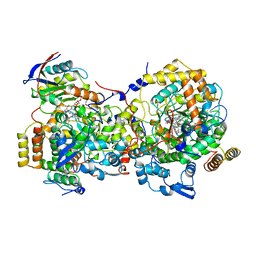 | | DIOL DEHYDRATASE-CYANOCOBALAMIN COMPLEX FROM KLEBSIELLA OXYTOCA | | 分子名称: | COBALAMIN, POTASSIUM ION, PROTEIN (DIOL DEHYDRATASE), ... | | 著者 | Shibata, N, Masuda, J, Tobimatsu, T, Toraya, T, Suto, K, Morimoto, Y, Yasuoka, N. | | 登録日 | 1999-01-27 | | 公開日 | 2000-01-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: X-ray structure of diol dehydratase.
Structure Fold.Des., 7, 1999
|
|
6J1C
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, off-state | | 分子名称: | CHLORIDE ION, GLYCEROL, Green fluorescent protein | | 著者 | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | 登録日 | 2018-12-28 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1B
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, on-state | | 分子名称: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | 著者 | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | 登録日 | 2018-12-28 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1A
 
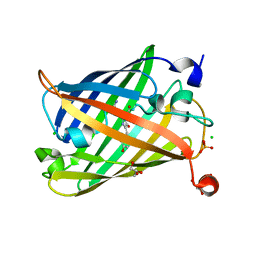 | | Photoswitchable fluorescent protein Gamillus, off-state | | 分子名称: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | 著者 | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | 登録日 | 2018-12-28 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6JXF
 
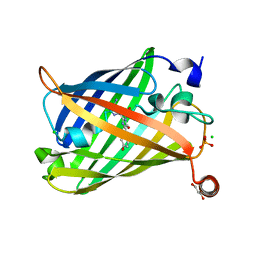 | | Photoswitchable fluorescent protein Gamillus, off-state (pH7.0) | | 分子名称: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | 著者 | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | 登録日 | 2019-04-23 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
7WAF
 
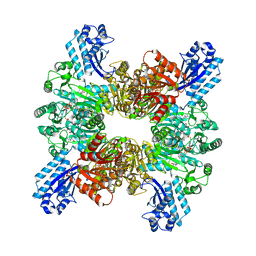 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS and 4x(beta-Asp-Arg) | | 分子名称: | 4x(beta-Asp-Arg), ARGININE, Cyanophycin synthase, ... | | 著者 | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | 登録日 | 2021-12-14 | | 公開日 | 2022-09-07 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (2.52 Å) | | 主引用文献 | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7WAC
 
 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) | | 分子名称: | Cyanophycin synthase | | 著者 | Kawasaki, M, Miyakawa, T, Yang, J, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | 登録日 | 2021-12-14 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
