5IZV
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - F222 | | 分子名称: | Uncharacterized protein RavZ | | 著者 | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | 登録日 | 2016-03-26 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.814 Å) | | 主引用文献 | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
3NGX
 
 | |
5ZYD
 
 | | Crystal Structure of Glucose Isomerase Soaked with Glucose | | 分子名称: | ACETATE ION, MAGNESIUM ION, Xylose isomerase | | 著者 | Nam, K.H. | | 登録日 | 2018-05-24 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural analysis of substrate recognition by glucose isomerase in Mn2+binding mode at M2 site in S. rubiginosus
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5ZYC
 
 | | Crystal Structure of Glucose Isomerase Soaked with Mn2+ | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | 著者 | Nam, K.H. | | 登録日 | 2018-05-24 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural analysis of substrate recognition by glucose isomerase in Mn2+binding mode at M2 site in S. rubiginosus
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5ZYE
 
 | |
5YCX
 
 | |
5YCS
 
 | |
5YCR
 
 | |
5YCV
 
 | |
5HZY
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - P6322 | | 分子名称: | Uncharacterized protein RavZ | | 著者 | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | 登録日 | 2016-02-03 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.548 Å) | | 主引用文献 | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
1FA2
 
 | | CRYSTAL STRUCTURE OF BETA-AMYLASE FROM SWEET POTATO | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-AMYLASE, alpha-D-glucopyranose-(1-4)-2-deoxy-beta-D-arabino-hexopyranose | | 著者 | Lee, B.I, Cheong, C.G, Suh, S.W. | | 登録日 | 2000-07-12 | | 公開日 | 2000-08-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystallization, molecular replacement solution, and refinement of tetrameric beta-amylase from sweet potato.
Proteins, 21, 1995
|
|
3A3F
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae,complexed with novel beta-lactam (FMZ) | | 分子名称: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxo-1-({(2R)-2-[(2-oxoimidazolidin-1-yl)amino]-2-phenylacetyl}amino)ethyl]-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | 著者 | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2009-06-12 | | 公開日 | 2009-12-22 | | 最終更新日 | 2013-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3I
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae, complexed with ampicillin (AIX) | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | 著者 | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2009-06-12 | | 公開日 | 2009-12-22 | | 最終更新日 | 2013-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3J
 
 | | Crystal structures of penicillin binding protein 5 from Haemophilus influenzae | | 分子名称: | PBP5, SULFATE ION | | 著者 | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2009-06-12 | | 公開日 | 2009-12-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3D
 
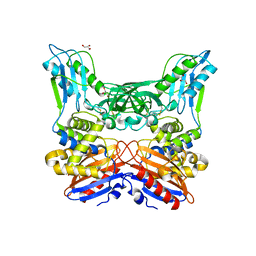 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae | | 分子名称: | GLYCEROL, Penicillin-binding protein 4 | | 著者 | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2009-06-12 | | 公開日 | 2009-12-22 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3E
 
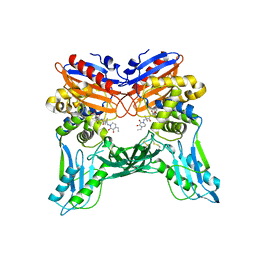 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae, complexed with novel beta-lactam (CMV) | | 分子名称: | (2R,4S)-2-[(1R)-1-({(2R)-2-[(4-ethyl-2,3-dioxopiperazin-1-yl)amino]-2-phenylacetyl}amino)-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | 著者 | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2009-06-12 | | 公開日 | 2009-12-22 | | 最終更新日 | 2013-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
1CNS
 
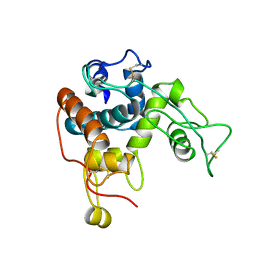 | |
8GUW
 
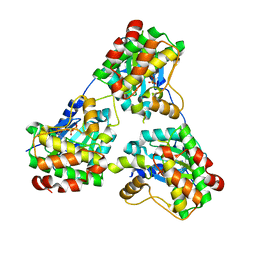 | |
4PQQ
 
 | | The crystal structure of discoidin domain from muskelin | | 分子名称: | Muskelin, PHOSPHATE ION, TETRAETHYLENE GLYCOL | | 著者 | Kim, K.-H, Hong, S.K, Kim, E.E. | | 登録日 | 2014-03-04 | | 公開日 | 2014-11-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure of mouse muskelin discoidin domain and biochemical characterization of its self-association.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4R3Z
 
 | | Crystal structure of human ArgRS-GlnRS-AIMP1 complex | | 分子名称: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1, Arginine--tRNA ligase, cytoplasmic, ... | | 著者 | Fu, Y, Kim, Y, Cho, Y. | | 登録日 | 2014-08-18 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (4.033 Å) | | 主引用文献 | Structure of the ArgRS-GlnRS-AIMP1 complex and its implications for mammalian translation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6LDK
 
 | |
2OHV
 
 | |
2OHG
 
 | |
2OHO
 
 | |
1M2K
 
 | | Sir2 homologue F159A mutant-ADP ribose complex | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | 著者 | Chang, J, Cho, Y. | | 登録日 | 2002-06-24 | | 公開日 | 2003-04-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
