7B3W
 
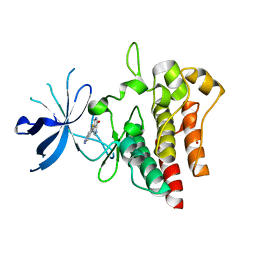 | | Crystal structure of c-MET bound by compound 4 | | 分子名称: | 1,2-ETHANEDIOL, 3-(6-fluoranyl-1~{H}-indazol-4-yl)-4,5-dihydro-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, Hepatocyte growth factor receptor | | 著者 | Collie, G.W. | | 登録日 | 2020-12-01 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3Z
 
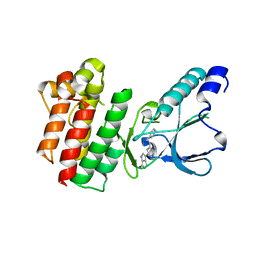 | | Crystal structure of c-MET bound by compound 5 | | 分子名称: | 3-[3-(phenylmethyl)-1~{H}-pyrrolo[2,3-b]pyridin-5-yl]-4,5-dihydro-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, DIMETHYL SULFOXIDE, Hepatocyte growth factor receptor | | 著者 | Collie, G.W. | | 登録日 | 2020-12-01 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3V
 
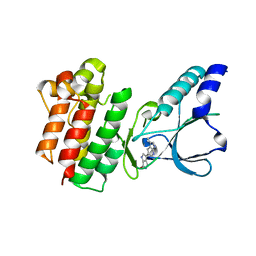 | | Crystal structure of c-MET bound by compound 3 | | 分子名称: | 3-(3-methyl-1~{H}-pyrrolo[2,3-b]pyridin-5-yl)-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, Hepatocyte growth factor receptor | | 著者 | Collie, G.W. | | 登録日 | 2020-12-01 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B44
 
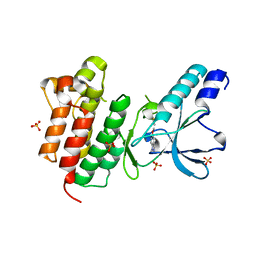 | | Crystal structure of c-MET bound by compound S1 | | 分子名称: | 5-methoxy-1~{H}-indazole, DIMETHYL SULFOXIDE, Hepatocyte growth factor receptor, ... | | 著者 | Collie, G.W. | | 登録日 | 2020-12-02 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
6JIT
 
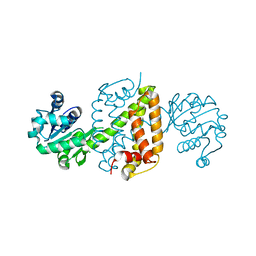 | | Complex structure of an imine reductase at 2.05 Angstrom resolution | | 分子名称: | 1-(2-phenylethyl)-3,4-dihydroisoquinoline, 6-phosphogluconate dehydrogenase NAD-binding protein, CHLORIDE ION, ... | | 著者 | Li, H, Wu, L, Zheng, G.W, Zhou, J.H. | | 登録日 | 2019-02-23 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.052 Å) | | 主引用文献 | Complex structure of an imine reductase at 2.05 Angstrom resolution
To Be Published
|
|
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | 分子名称: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | 登録日 | 2022-04-19 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
1KQQ
 
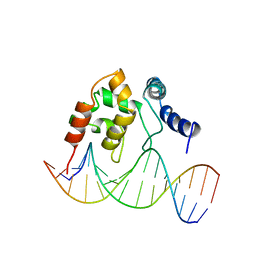 | | Solution Structure of the Dead ringer ARID-DNA Complex | | 分子名称: | 5'-D(*CP*CP*AP*CP*AP*TP*CP*AP*AP*TP*AP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*TP*AP*TP*TP*GP*AP*TP*GP*TP*GP*G)-3', DEAD RINGER PROTEIN | | 著者 | Iwahara, J, Iwahara, M, Daughdrill, G.W, Ford, J, Clubb, R.T. | | 登録日 | 2002-01-07 | | 公開日 | 2002-03-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA.
EMBO J., 21, 2002
|
|
1MO7
 
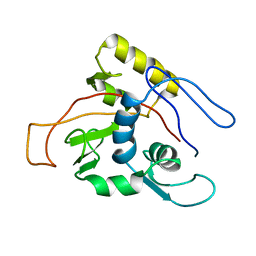 | | ATPase | | 分子名称: | Sodium/Potassium-transporting ATPase alpha-1 chain | | 著者 | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | 登録日 | 2002-09-08 | | 公開日 | 2003-06-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
1MO8
 
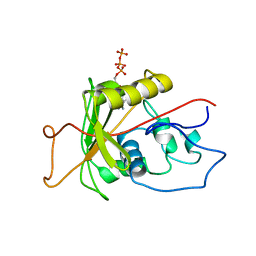 | | ATPase | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Sodium/Potassium-Transporting ATPase alpha-1 | | 著者 | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | 登録日 | 2002-09-08 | | 公開日 | 2003-06-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
1MZ8
 
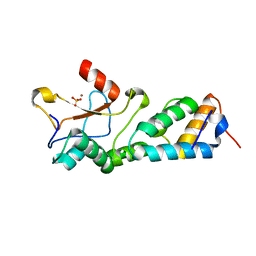 | | CRYSTAL STRUCTURES OF THE NUCLEASE DOMAIN OF COLE7/IM7 IN COMPLEX WITH A PHOSPHATE ION AND A ZINC ION | | 分子名称: | Colicin E7, Colicin E7 immunity protein, PHOSPHATE ION, ... | | 著者 | Sui, M.J, Tsai, L.C, Hsia, K.C, Doudeva, L.G, Ku, W.Y, Han, G.W, Yuan, H.S. | | 登録日 | 2002-10-07 | | 公開日 | 2002-12-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Metal ions and phosphate binding in the H-N-H motif: crystal structures of the nuclease domain of ColE7/Im7 in complex with a phosphate ion and different divalent metal ions
PROTEIN SCI., 11, 2002
|
|
1G4Q
 
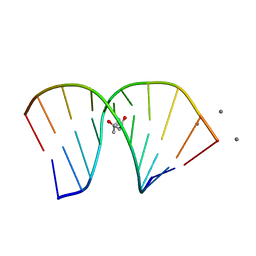 | | RNA/DNA HYBRID DECAMER OF CAAAGAAAAG/CTTTTCTTTG | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', ... | | 著者 | Han, G.W. | | 登録日 | 2000-10-27 | | 公開日 | 2003-07-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Direct-methods determination of an RNA/DNA hybrid decamer at 1.15 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2GPN
 
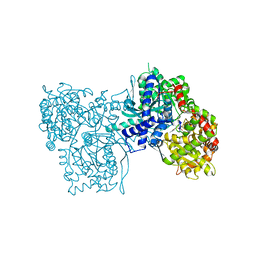 | | 100 K STRUCTURE OF GLYCOGEN PHOSPHORYLASE AT 2.0 ANGSTROMS RESOLUTION | | 分子名称: | GLYCOGEN PHOSPHORYLASE B | | 著者 | Gregoriou, M, Noble, M.E.M, Watson, K.A, Garman, E.F, Krulle, T.M, De La Fuente, C, Fleet, G.W.J, Oikonomakos, N.G, Johnson, L.N. | | 登録日 | 1998-03-26 | | 公開日 | 1998-07-01 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 A resolution and 100 K: the role of the water structure and its contribution to binding.
Protein Sci., 7, 1998
|
|
7TBM
 
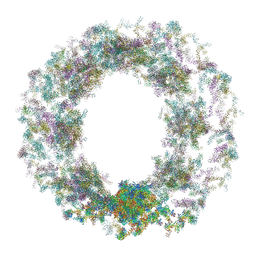 | | Composite structure of the dilated human nuclear pore complex (NPC) generated with a 37A in situ cryo-ET map of CD4+ T cell NPC | | 分子名称: | DDX19, NUP107 CTD, NUP107 NTD, ... | | 著者 | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (37 Å) | | 主引用文献 | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7TBK
 
 | | Composite structure of the dilated human nuclear pore complex (NPC) symmetric core generated with a 37A in situ cryo-ET map of CD4+ T cell NPC | | 分子名称: | NUP107 CTD, NUP107 NTD, NUP133, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (37 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7TBI
 
 | | Composite structure of the S. cerevisiae nuclear pore complex (NPC) | | 分子名称: | Dyn2, Nic96 R1, Nic96 R2, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (25 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7TBJ
 
 | | Composite structure of the human nuclear pore complex (NPC) symmetric core generated with a 12A cryo-ET map of the purified HeLa cell NPC | | 分子名称: | NUP107 CTD, NUP107 NTD, NUP133, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2022-06-29 | | 実験手法 | ELECTRON MICROSCOPY (23 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7TBL
 
 | | Composite structure of the human nuclear pore complex (NPC) cytoplasmic face generated with a 12A cryo-ET map of the purified HeLa cell NPC | | 分子名称: | DDX19, ELYS, GLE1, ... | | 著者 | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (23 Å) | | 主引用文献 | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7SUS
 
 | | Crystal structure of Apelin receptor in complex with small molecule | | 分子名称: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor, ... | | 著者 | Xu, F, Yue, Y, Liu, L.E, Han, G.W, Hanson, M. | | 登録日 | 2021-11-18 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5F4N
 
 | | Multi-parameter lead optimization to give an oral CHK1 inhibitor clinical candidate: (R)-5-((4-((morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737) | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase Chk1, ... | | 著者 | Collins, I, Garrett, M.D, van Montfort, R, Osborne, J.D, Matthews, T.P, McHardy, T, Proisy, N, Cheung, K.J, Lainchbury, M, Brown, N, Walton, M.I, Eve, P.D, Boxall, K.J, Hayes, A, Henley, A.T, Valenti, M.R, De Haven Brandon, A.K, Box, G, Westwood, I.M, Jamin, Y, Robinson, S.P, Leonard, P, Reader, J.C, Aherne, G.W, Raynaud, F.I, Eccles, S.A. | | 登録日 | 2015-12-03 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Multiparameter Lead Optimization to Give an Oral Checkpoint Kinase 1 (CHK1) Inhibitor Clinical Candidate: (R)-5-((4-((Morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737).
J.Med.Chem., 59, 2016
|
|
7TH8
 
 | | Chickpea (Cicer arientinum) nodule-specific cysteine-rich peptide NCR13: Solution NMR structure of the isomer with C4:C10, C15:C30, and C23:C28 disulfide bonds | | 分子名称: | Nodule cysteine-rich protein 13 | | 著者 | Buchko, G.W, Zhou, M, Shah, D.M, Velivelli, S.L.S. | | 登録日 | 2022-01-10 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Chickpea (Cicer arientinum) nodule-specific cysteine-rich peptide NCR13: Solution NMR structure of the isomer with C4:C10, C15:C30, and C23:C28 disulfide bonds
To Be Published
|
|
7TOD
 
 | | Solution structure of the phosphatidylinositol 3-phosphate binding domain from the Legionella effector SetA | | 分子名称: | Subversion of eukaryotic traffic protein A | | 著者 | Beck, W.H.J, Enoki, T.A, Feigenson, G.W, Nicholson, L, Oswald, R, Mao, Y. | | 登録日 | 2022-01-24 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the phosphatidylinositol 3-phosphate binding domain from the Legionella effector SetA
To Be Published
|
|
7TXX
 
 | |
5ET9
 
 | | Racemic crystal structures of Pribnow box consensus promoter sequence (P21/n) | | 分子名称: | BARIUM ION, Pribnow box consensus sequence- template strand, Pribnow box non-template strand | | 著者 | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | 登録日 | 2015-11-17 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
7TVP
 
 | | Viral AMG chitosanase V-Csn, E157Q mutant, chitotriose complex | | 分子名称: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, GLYCEROL, Viral chitosanase V-Csn E157Q mutant chitotriose complex | | 著者 | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | 登録日 | 2022-02-05 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVM
 
 | | Viral AMG chitosanase V-Csn, apo structure, crystal form 2 | | 分子名称: | 1,2-ETHANEDIOL, Viral chitosanase V-Csn | | 著者 | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | 登録日 | 2022-02-05 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
