8VNO
 
 | |
8VLC
 
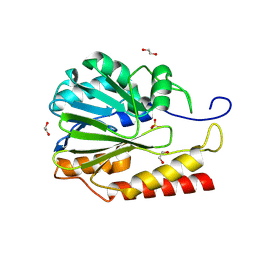 | | Crystal structure of Zn-dependent hydrolase from Salmonella typhimurium LT2 | | 分子名称: | 1,2-ETHANEDIOL, HARLDQ motif MBL-fold protein, SULFATE ION | | 著者 | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2024-01-11 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of Zn-dependent hydrolase from Salmonella typhimurium LT2
To Be Published
|
|
8VMW
 
 | |
8VN7
 
 | |
8VNH
 
 | |
8VNT
 
 | |
8VMV
 
 | |
8VN1
 
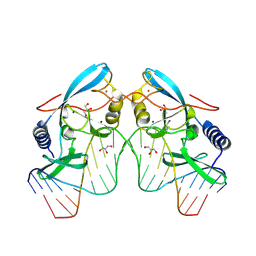 | |
8VNB
 
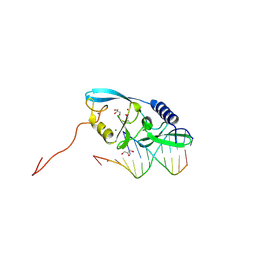 | |
8VNL
 
 | |
1BXC
 
 | |
1BXB
 
 | |
5BS6
 
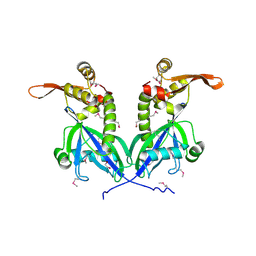 | | Apo structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI | | 分子名称: | 1,2-ETHANEDIOL, transcriptional regulator AraR | | 著者 | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-06-01 | | 公開日 | 2015-06-17 | | 最終更新日 | 2015-12-16 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5DD4
 
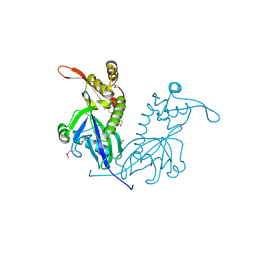 | | Apo structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, TRANSCRIPTIONAL REGULATOR AraR | | 著者 | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-08-24 | | 公開日 | 2015-09-09 | | 最終更新日 | 2015-12-16 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5C4Y
 
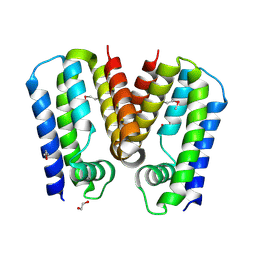 | | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes | | 分子名称: | 1,2-ETHANEDIOL, Putative transcription regulator Lmo0852 | | 著者 | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-06-18 | | 公開日 | 2015-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes
to be published
|
|
5DDG
 
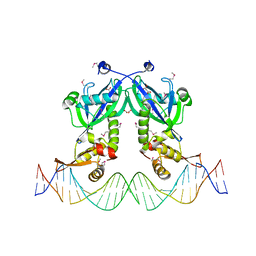 | | The structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI in complex with target double strand DNA | | 分子名称: | DNA (27-MER), FORMIC ACID, MALONIC ACID, ... | | 著者 | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-08-24 | | 公開日 | 2015-09-09 | | 最終更新日 | 2015-12-16 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5DEQ
 
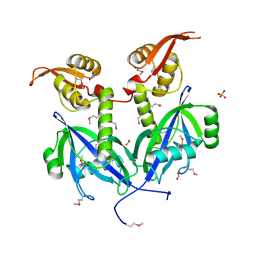 | | Crystal structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI in complex with L-arabinose | | 分子名称: | FORMIC ACID, SULFATE ION, TRANSCRIPTIONAL REGULATOR AraR, ... | | 著者 | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-08-25 | | 公開日 | 2015-10-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
7LZG
 
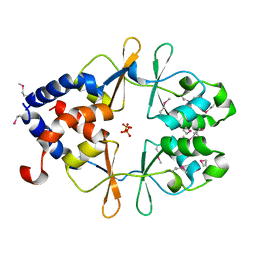 | |
7M92
 
 | |
7MH7
 
 | |
5E2C
 
 | | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv | | 分子名称: | Xaa-Pro dipeptidase | | 著者 | Chang, C, Endres, L, Endres, M, SACCHETTINI, J, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2015-09-30 | | 公開日 | 2015-10-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv
To Be Published
|
|
5ER3
 
 | | Crystal structure of ABC transporter system solute-binding protein from Rhodopirellula baltica SH 1 | | 分子名称: | CALCIUM ION, GLYCEROL, Sugar ABC transporter, ... | | 著者 | Chang, C, Duke, N, Endres, M, Mack, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-11-13 | | 公開日 | 2015-11-25 | | 実験手法 | X-RAY DIFFRACTION (2.105 Å) | | 主引用文献 | Crystal structure of ABC transporter system solute-binding protein from Rhodopirellula baltica SH 1
To Be Published
|
|
5EVH
 
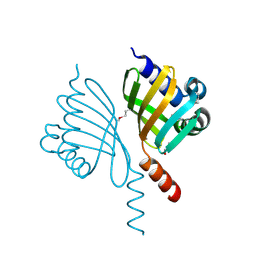 | | Crystal structure of known function protein from Kribbella flavida DSM 17836 | | 分子名称: | GLYCEROL, Uncharacterized protein | | 著者 | Chang, C, Duke, N, Endres, M, Chhor, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-11-19 | | 公開日 | 2015-12-02 | | 実験手法 | X-RAY DIFFRACTION (1.852 Å) | | 主引用文献 | Crystal structure of known function protein from Kribbella flavida DSM 17836
To Be Published
|
|
5FDA
 
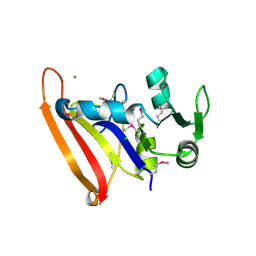 | | The high resolution structure of apo form dihydrofolate reductase from Yersinia pestis at 1.55 A | | 分子名称: | CHLORIDE ION, Dihydrofolate reductase | | 著者 | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-12-15 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.549 Å) | | 主引用文献 | structure of dihydrofolate reductase from Yersinia pestis complex with
To Be Published
|
|
6NBK
 
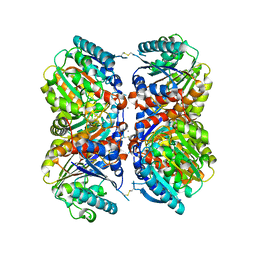 | | Crystal structure of Arginase from Bacillus cereus | | 分子名称: | Arginase, CALCIUM ION, MANGANESE (II) ION | | 著者 | Chang, C, Evdokimova, E, Mcchesney, M, Joachimiak, A, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-12-07 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Crystal structure of Arginase from Bacillus cereus
To Be Published
|
|
