5WE1
 
 | | Structural Basis for Shelterin Bridge Assembly | | 分子名称: | Protection of telomeres protein poz1,Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ZINC ION | | 著者 | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | 登録日 | 2017-07-06 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.202 Å) | | 主引用文献 | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
5WE0
 
 | | Structural Basis for Shelterin Bridge Assembly | | 分子名称: | DNA-binding protein rap1, Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ... | | 著者 | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | 登録日 | 2017-07-06 | | 公開日 | 2017-12-20 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
5WE2
 
 | | Structural Basis for Telomere Length Regulation by the Shelterin Bridge | | 分子名称: | DNA-binding protein rap1, Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ... | | 著者 | Kim, J.-K, Liu, J, Hu, X, Sankaran, B, Qiao, F. | | 登録日 | 2017-07-06 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
4PM5
 
 | | Crystal structure of CTX-M-14 S70G beta-lactamase in complex with cefotaxime at 1.26 Angstroms resolution | | 分子名称: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14, SULFATE ION | | 著者 | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | 登録日 | 2014-05-20 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.261 Å) | | 主引用文献 | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
8SQF
 
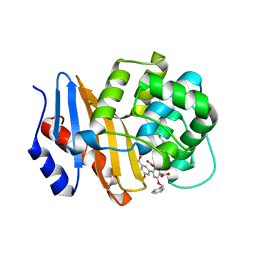 | | OXA-48 bound to inhibitor CDD-2725 | | 分子名称: | (1M)-3'-(benzyloxy)-5-hydroxy[1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | 著者 | Park, S, Judge, A, Fan, J, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2023-05-04 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
8SQG
 
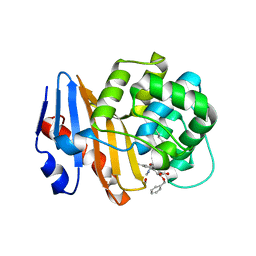 | | OXA-48 bound to inhibitor CDD-2801 | | 分子名称: | (1M)-3'-(benzyloxy)-5-[2-(methylamino)-2-oxoethoxy][1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | 著者 | Park, S, Judge, A, Fan, J, Sankaran, B, Palzkill, T. | | 登録日 | 2023-05-04 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
8SJ3
 
 | |
8T6C
 
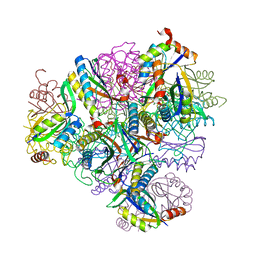 | | Crystal structure of T33-18.2: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | 分子名称: | T33-18.2 : A, T33-18.2 : B | | 著者 | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | 登録日 | 2023-06-15 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8T6N
 
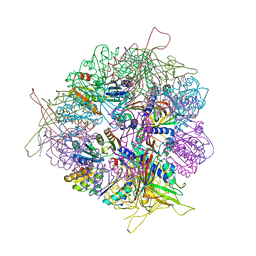 | | Crystal structure of T33-27.1: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | 分子名称: | T33-27.1 : A, T33-27.1 : B | | 著者 | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | 登録日 | 2023-06-16 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (3.63 Å) | | 主引用文献 | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8T6E
 
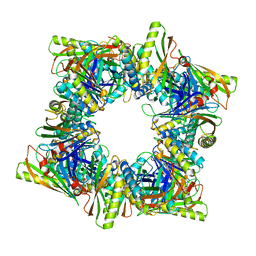 | | Crystal structure of T33-28.3: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | 分子名称: | T33-28.3: A, T33-28.3: B | | 著者 | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | 登録日 | 2023-06-15 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4P12
 
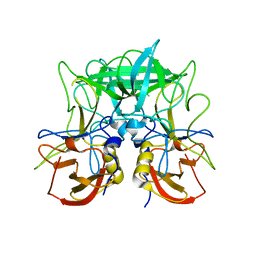 | | Native Structure of the P domain from a GI.7 Norovirus variant. | | 分子名称: | Major capsid protein | | 著者 | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | 登録日 | 2014-02-24 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6011 Å) | | 主引用文献 | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P25
 
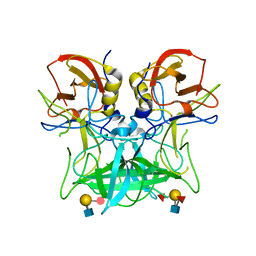 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeY HBGA. | | 分子名称: | Major capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | 登録日 | 2014-03-01 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.4991 Å) | | 主引用文献 | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P26
 
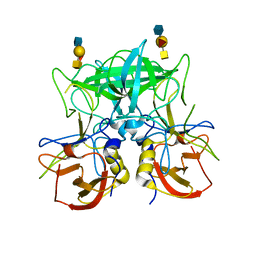 | | Structure of the P domain from a GI.7 Norovirus variant in complex with A-type 2 HBGA | | 分子名称: | P domain of VP1, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | 登録日 | 2014-03-01 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P2N
 
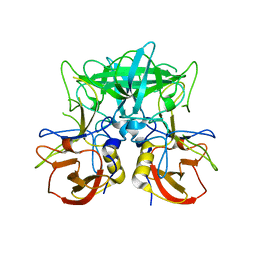 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeX HBGA | | 分子名称: | Major capsid protein, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | 登録日 | 2014-03-04 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4PM8
 
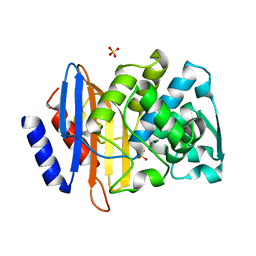 | | Crystal structure of CTX-M-14 S70G:S237A beta-lactamase at 1.17 Angstroms resolution | | 分子名称: | Beta-lactamase CTX-M-14, SULFATE ION | | 著者 | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | 登録日 | 2014-05-20 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.169 Å) | | 主引用文献 | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4P1V
 
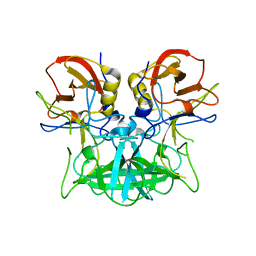 | | Structure of the P domain from a GI.7 Norovirus variant in complex with H-type 2 HBGA | | 分子名称: | P domain of VPI, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | 登録日 | 2014-02-27 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5497 Å) | | 主引用文献 | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4PMA
 
 | | Crystal structure of CTX-M-14 S70G:S237A:R276A beta-lactamase at 1.39 Angstroms resolution | | 分子名称: | Beta-lactamase CTX-M-14, SULFATE ION | | 著者 | Cardenas, A.M, Adamski, C.J, Brown, N.G, Horton, L.B, Sankaran, B, Palzkill, T. | | 登録日 | 2014-05-20 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.399 Å) | | 主引用文献 | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4P3I
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeA HBGA. | | 分子名称: | P domain of VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | 登録日 | 2014-03-07 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6941 Å) | | 主引用文献 | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4PM9
 
 | | Crystal structure of CTX-M-14 S70G:S237A:R276A beta-lactamase in complex with cefotaxime at 1.45 Angstroms resolution | | 分子名称: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14 | | 著者 | Cardenas, A.M, Adamski, C.J, Sankaran, B, Brown, N.G, Horton, L.B, Palzkill, T. | | 登録日 | 2014-05-20 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.454 Å) | | 主引用文献 | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4PM7
 
 | | Crystal structure of CTX-M-14 S70G:S237A in complex with cefotaxime at 1.29 Angstroms resolution | | 分子名称: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14 | | 著者 | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | 登録日 | 2014-05-20 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4PM6
 
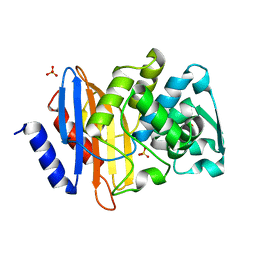 | | Crystal structure of CTX-M-14 S70G beta-lactamase at 1.56 Angstroms resolution | | 分子名称: | Beta-lactamase CTX-M-14, SULFATE ION | | 著者 | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | 登録日 | 2014-05-20 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4QX5
 
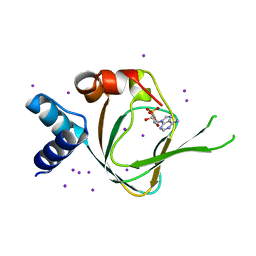 | | Neutron diffraction reveals hydrogen bonds critical for cGMP-selective activation: Insights for PKG agonist design | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, IODIDE ION, cGMP-dependent protein kinase 1 | | 著者 | Huang, G.Y, Gerlits, O.O, Blakeley, M.P, Sankaran, B, Kovalevsky, A.Y, Kim, C. | | 登録日 | 2014-07-18 | | 公開日 | 2014-11-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.318 Å) | | 主引用文献 | Neutron Diffraction Reveals Hydrogen Bonds Critical for cGMP-Selective Activation: Insights for cGMP-Dependent Protein Kinase Agonist Design.
Biochemistry, 53, 2014
|
|
3MYU
 
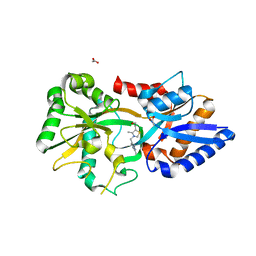 | | Mycoplasma genitalium MG289 | | 分子名称: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, ACETATE ION, High affinity transport system protein p37 | | 著者 | Sippel, K.H, Boehlein, S.K, Govindasamy, L, Namiki, K, Satai, Y, Quirit, J.G, Agbandje-McKenna, M, Goodison, S, Rosser, C.J, Sankaran, B, McKenna, R. | | 登録日 | 2010-05-11 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Insights into Mycoplasma genitalium metabolism revealed by the structure of MG289, an extracytoplasmic thiamine binding lipoprotein.
Proteins, 79, 2011
|
|
4RVA
 
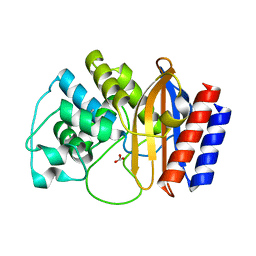 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for deacylation | | 分子名称: | BICARBONATE ION, Beta-lactamase TEM | | 著者 | Stojanoski, V, Chow, D.-C, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2014-11-25 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4397 Å) | | 主引用文献 | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
4RX3
 
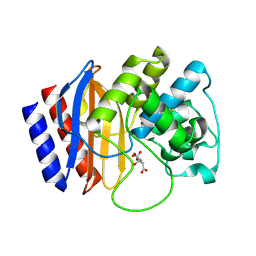 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for catalysis | | 分子名称: | Beta-lactamase TEM, CITRATE ANION | | 著者 | Stojanoski, V, Chow, D, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2014-12-08 | | 公開日 | 2015-03-04 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
