7ENF
 
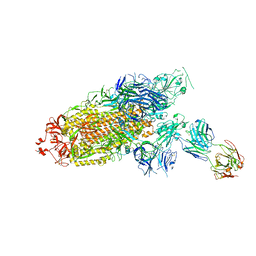 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab30, ... | | 著者 | Wang, X.F, Zhu, Y.Q. | | 登録日 | 2021-04-16 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENG
 
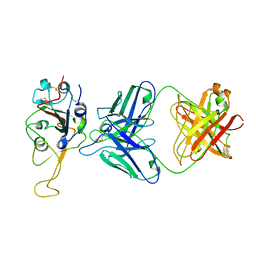 | |
7F46
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | 著者 | Wang, X.F, Zhu, Y.Q. | | 登録日 | 2021-06-17 | | 公開日 | 2022-03-23 | | 最終更新日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (4.79 Å) | | 主引用文献 | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
2FKC
 
 | |
2FLC
 
 | |
2FKH
 
 | |
2FL3
 
 | |
8XIR
 
 | | Structure of pasireotide-SSTR3 G protein complex | | 分子名称: | G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | 登録日 | 2023-12-19 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.52 Å) | | 主引用文献 | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3
To Be Published
|
|
8XIO
 
 | | Structure of L797591-SSTR1 G protein complex | | 分子名称: | (2~{S})-~{N}-[(4~{S})-6-azanyl-2,2,4-trimethyl-hexyl]-3-naphthalen-1-yl-2-[[2-phenylethyl(2-pyridin-2-ylethyl)carbamoyl]amino]propanamide, G-alpha-i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | 登録日 | 2023-12-19 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.65 Å) | | 主引用文献 | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3
To Be Published
|
|
8XIQ
 
 | | Structure of L796778-SSTR3 G protein complex | | 分子名称: | G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | 登録日 | 2023-12-19 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3
To Be Published
|
|
8XIP
 
 | | Structure of Pasireotide-SSTR1 G protein complex | | 分子名称: | 004-DTR-LYS-TY5-PHE-A1D5E, G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | 登録日 | 2023-12-19 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3
To Be Published
|
|
1MVH
 
 | | structure of the SET domain histone lysine methyltransferase Clr4 | | 分子名称: | Cryptic loci regulator 4, NICKEL (II) ION, SULFATE ION, ... | | 著者 | Min, J.R, Zhang, X, Cheng, X.D, Grewal, S.I.S, Xu, R.-M. | | 登録日 | 2002-09-25 | | 公開日 | 2002-10-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the SET domain histone lysine methyltransferase Clr4.
Nat.Struct.Biol., 9, 2002
|
|
7DAJ
 
 | | The crystal structure of serotonin N-acetyltransferase in complex with acetyl-CoA from Oryza Sativa | | 分子名称: | ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, chloroplastic | | 著者 | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | 登録日 | 2020-10-16 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAI
 
 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa | | 分子名称: | Serotonin N-acetyltransferase 1, chloroplastic | | 著者 | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | 登録日 | 2020-10-16 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAL
 
 | | The crystal structure of a serotonin N-acetyltransferase in complex with serotonin and acetyl-CoA from Oryza Sativa | | 分子名称: | ACETYL COENZYME *A, SEROTONIN, Serotonin N-acetyltransferase 1, ... | | 著者 | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | 登録日 | 2020-10-16 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAK
 
 | | The crystal structure of a serotonin N-acetyltransferase in complex with 5-Methoxytryptamine and acetyl-CoA from Oryza Sativa | | 分子名称: | 2-(5-methoxy-1H-indol-3-yl)ethanamine, ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, ... | | 著者 | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | 登録日 | 2020-10-16 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL0
 
 | | The mutant E310G/A314Y of 3,5-DAHDHcca complex with NADPH | | 分子名称: | 1,2-ETHANEDIOL, 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, ... | | 著者 | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | 登録日 | 2020-11-25 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL7
 
 | | The wild-type structure of 3,5-DAHDHcca | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,5-diaminohexanoate dehydrogenase, ... | | 著者 | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | 登録日 | 2020-11-26 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.30065823 Å) | | 主引用文献 | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL1
 
 | | The mutant E310G/G323S structure of 3,5-DAHDHcca complex with NADPH | | 分子名称: | 3,5-diaminohexanoate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | 登録日 | 2020-11-25 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL3
 
 | | The structure of 3,5-DAHDHcca complex with NADPH | | 分子名称: | 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | 登録日 | 2020-11-25 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.84606934 Å) | | 主引用文献 | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DGB
 
 | |
7DGH
 
 | |
7DGF
 
 | |
7DHJ
 
 | | The co-crystal structure of SARS-CoV-2 main protease with the peptidomimetic inhibitor (S)-2-cinnamamido-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)pent-4-ynamide | | 分子名称: | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide, 3C-like proteinase | | 著者 | Shang, L.Q, Wang, H, Deng, W.L, Xing, S, Wang, Y.X. | | 登録日 | 2020-11-15 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.962 Å) | | 主引用文献 | The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Eur.J.Med.Chem., 238, 2022
|
|
7DGG
 
 | |
