9B2C
 
 | |
4Q12
 
 | |
4PY3
 
 | |
4F4F
 
 | |
9C6O
 
 | |
9DAK
 
 | |
9E0I
 
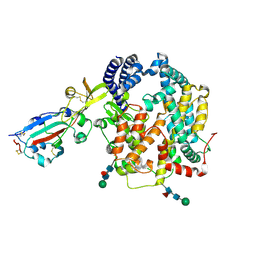 | |
2MJ3
 
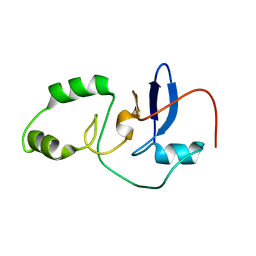 | |
6C0E
 
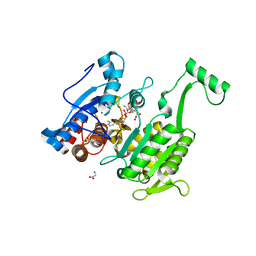 | | Crystal Structure of Isocitrate Dehydrogenase from Legionella pneumophila with bound NADPH with an alpha-ketoglutarate adduct | | 分子名称: | (3~{S})-3-[(4~{S})-3-aminocarbonyl-1-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]-4~{H}-pyridin-4-yl]-2-oxidanylidene-pentanedioic acid, CHLORIDE ION, GLYCINE, ... | | 著者 | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2017-12-29 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of Isocitrate Dehydrogenase from Legionella pneumophila with bound NADPH with an ??-ketoglutarate adduct
to be published
|
|
8VYE
 
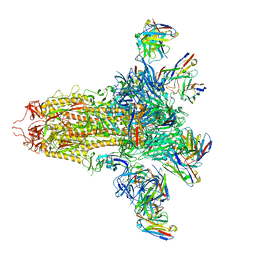 | | SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, S2L20 Light Chain, ... | | 著者 | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2024-02-08 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
8VYF
 
 | | SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, ... | | 著者 | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2024-02-08 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
8W22
 
 | | Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF of UmbC1 and UmbA1) | | 分子名称: | Intein C-terminal splicing domain-containing protein, Secreted esterase, Secreted protein | | 著者 | Park, Y.J, Zhao, Q, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | 登録日 | 2024-02-19 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Streptomyces umbrella toxin particles block hyphal growth of competing species.
Nature, 629, 2024
|
|
8W20
 
 | | Umb1 umbrella toxin particle | | 分子名称: | Intein C-terminal splicing domain-containing protein, Secreted esterase, Secreted protein | | 著者 | Park, Y.J, Zhao, Q, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | 登録日 | 2024-02-19 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Streptomyces umbrella toxin particles block hyphal growth of competing species.
Nature, 629, 2024
|
|
8VWP
 
 | |
8VYG
 
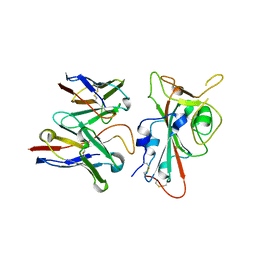 | | SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Heavy Chain, S309 Light Chain, ... | | 著者 | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2024-02-08 | | 公開日 | 2024-03-27 | | 最終更新日 | 2025-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
9DGO
 
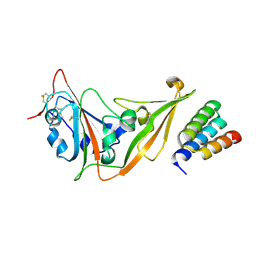 | |
4EFI
 
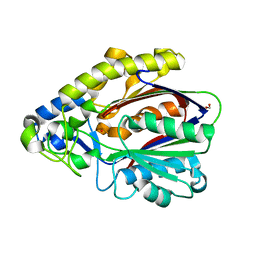 | | Crystal Structure of 3-oxoacyl-(Acyl-carrier protein) Synthase from Burkholderia Xenovorans LB400 | | 分子名称: | 3-oxoacyl-(Acyl-carrier protein) synthase, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Seattle Structural Genomics Center for Infectious Disease, Craig, T.K, Abendroth, J, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2012-03-29 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
6E54
 
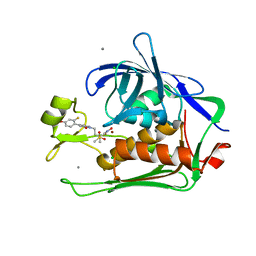 | |
8U29
 
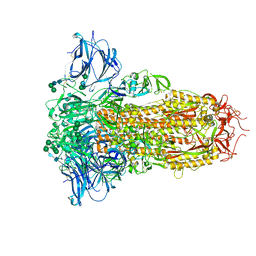 | | Prefusion structure of the PRD-0038 spike glycoprotein ectodomain trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PRD-0038 Spike glycoprotein, ... | | 著者 | Lee, J, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2023-09-05 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Broad receptor tropism and immunogenicity of a clade 3 sarbecovirus.
Cell Host Microbe, 31, 2023
|
|
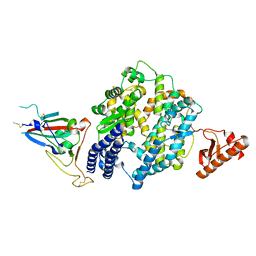
8U0T
 
 | |
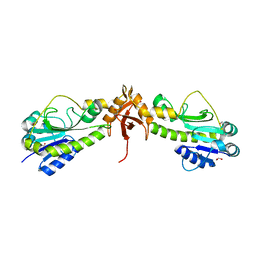
4PZU
 
 | |
4QTP
 
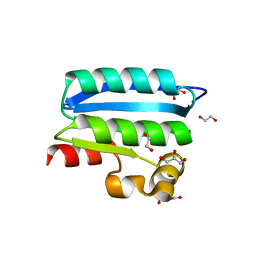 | | Crystal Structure of an Anti-sigma Factor Antagonist from Mycobacterium paratuberculosis | | 分子名称: | 1,2-ETHANEDIOL, Anti-sigma factor antagonist, CITRIC ACID, ... | | 著者 | Dranow, D.M, Clifton, M.C, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2014-07-08 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of an Anti-sigma Factor Antagonist from Mycobacterium paratuberculosis
TO BE PUBLISHED
|
|
6BLA
 
 | | Structure of AMM01 Fab, an anti EBV gH/gL neutralizing antibody | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AMM01 Fab Heavy chain, ... | | 著者 | Pancera, M, Weidle, C, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2017-11-09 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus.
Immunity, 48, 2018
|
|
3CEZ
 
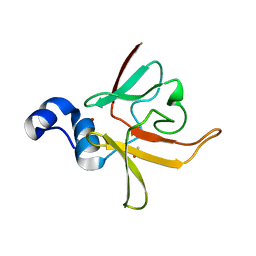 | | Crystal structure of methionine-R-sulfoxide reductase from Burkholderia pseudomallei | | 分子名称: | ACETIC ACID, Methionine-R-sulfoxide reductase, ZINC ION | | 著者 | Staker, B, Napuli, A, Nakazawa, S.H, Castaneda, L, Alkafeef, S, Vanvoorhis, W, Stewart, L, Myler, P, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2008-02-29 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Methionine-R-sulfoxide reductase from Burkholderia pseudomallei.
To be Published
|
|
8SOY
 
 | |
