7UCP
 
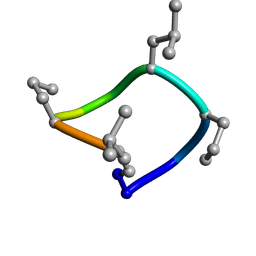 | | computationally designed macrocycle | | 分子名称: | computationally designed cyclic peptide D8.3.p2 | | 著者 | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | 登録日 | 2022-03-17 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
7UBC
 
 | |
7UBE
 
 | |
7UBG
 
 | |
4DEQ
 
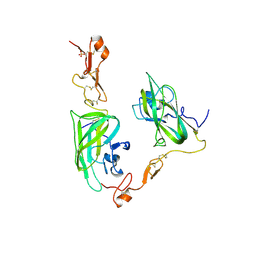 | | Structure of the Neuropilin-1/VEGF-A complex | | 分子名称: | Neuropilin-1, Vascular endothelial growth factor A, PHOSPHATE ION | | 著者 | Vander Kooi, C.W. | | 登録日 | 2012-01-21 | | 公開日 | 2012-02-08 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.649 Å) | | 主引用文献 | Structural Basis for Selective Vascular Endothelial Growth Factor-A (VEGF-A) Binding to Neuropilin-1.
J.Biol.Chem., 287, 2012
|
|
8ZF4
 
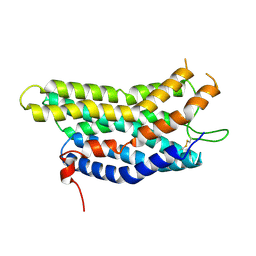 | |
8ZFB
 
 | |
8ZF7
 
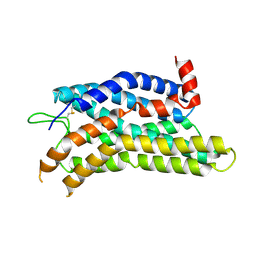 | |
8ZFC
 
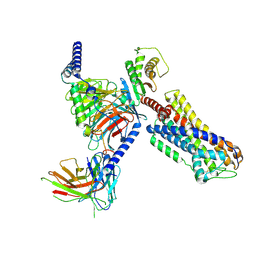 | | Cryo-EM structure of the mmGPR4-Gs complex in pH7.6 | | 分子名称: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wen, X, Rong, N.K, Yang, F, Sun, J.P. | | 登録日 | 2024-05-07 | | 公開日 | 2025-02-26 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZFE
 
 | |
8ZFA
 
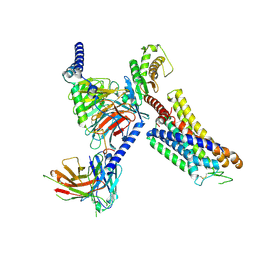 | | Cryo-EM structure of the xtGPR4-Gs complex in pH7.2 | | 分子名称: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | 登録日 | 2024-05-07 | | 公開日 | 2025-02-26 | | 実験手法 | ELECTRON MICROSCOPY (2.96 Å) | | 主引用文献 | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZF9
 
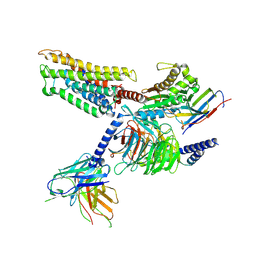 | | Cryo-EM structure of the mmGPR4-Gs complex in pH7.2 | | 分子名称: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wen, X, Rong, N.K, Yang, F, Sun, J.P. | | 登録日 | 2024-05-07 | | 公開日 | 2025-02-26 | | 実験手法 | ELECTRON MICROSCOPY (2.56 Å) | | 主引用文献 | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZD1
 
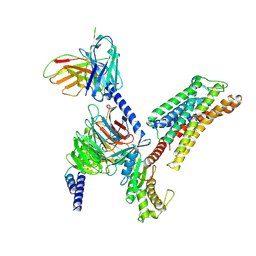 | | Cryo-EM structure of the xGPR4-Gs complex in pH6.2 | | 分子名称: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | 登録日 | 2024-04-30 | | 公開日 | 2025-02-26 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZF6
 
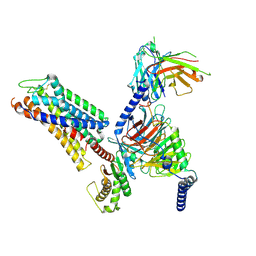 | | Cryo-EM structure of the xGPR4-Gs complex in pH6.7 | | 分子名称: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | 登録日 | 2024-05-07 | | 公開日 | 2025-02-26 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZFD
 
 | |
7KVV
 
 | |
7KVT
 
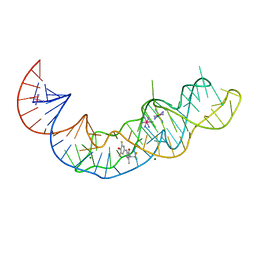 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T with iridium (III) ions | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | 著者 | Truong, L, Ferre-D'Amare, A.R. | | 登録日 | 2020-11-28 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
7KVU
 
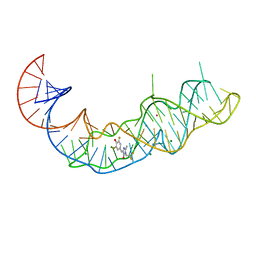 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | 著者 | Truong, L, Ferre-D'Amare, A.R. | | 登録日 | 2020-11-28 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
3N6M
 
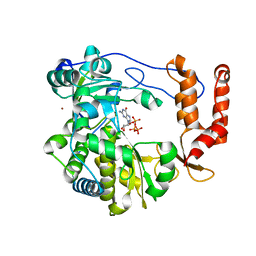 | | Crystal structure of EV71 RdRp in complex with GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, NICKEL (II) ION, RNA-dependent RNA polymerase | | 著者 | Wu, Y, Lou, Z.Y, Miao, Y, Yu, Y, Rao, Z.H. | | 登録日 | 2010-05-26 | | 公開日 | 2011-06-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
7MU0
 
 | | MtbEttA in the ADP bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Energy-dependent translational throttle protein EttA, MAGNESIUM ION | | 著者 | Cui, Z, Zhang, J. | | 登録日 | 2021-05-14 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
4GGL
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity | | 分子名称: | 7-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)pyrido[2,3-b]pyrazin-2(1H)-one, DNA gyrase subunit B, GLYCEROL | | 著者 | Bensen, D.C, Creighton, C.J, Tari, L.W. | | 登録日 | 2012-08-06 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3N6L
 
 | |
4GFN
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic | | 分子名称: | (1R,5S,6s)-3-[5-chloro-6-ethyl-2-(pyrimidin-5-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]-3-azabicyclo[3.1.0]hexan-6-amine, DNA gyrase subunit B, GLYCEROL | | 著者 | Bensen, D.C, Trzoss, M, Tari, L.W. | | 登録日 | 2012-08-03 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HCU
 
 | | Crystal structure of ITK in complext with compound 40 | | 分子名称: | 3-{4-amino-1-[(3R)-1-propanoylpiperidin-3-yl]-1H-pyrazolo[3,4-d]pyrimidin-3-yl}-N-[4-(propan-2-yl)phenyl]benzamide, Tyrosine-protein kinase ITK/TSK | | 著者 | Han, S, Caspers, N. | | 登録日 | 2012-10-01 | | 公開日 | 2012-11-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Covalent inhibitors of interleukin-2 inducible T cell kinase (itk) with nanomolar potency in a whole-blood assay.
J.Med.Chem., 55, 2012
|
|
4LWP
 
 | | Crystal structure of PRMT6-SAH | | 分子名称: | Arginine N-methyltransferase, putative, IODIDE ION, ... | | 著者 | Zhu, Y, Wang, C, Shi, Y, Teng, M. | | 登録日 | 2013-07-28 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.353 Å) | | 主引用文献 | Crystal Structure of Arginine Methyltransferase 6 from Trypanosoma brucei
Plos One, 9, 2014
|
|
