2KV8
 
 | | Solution structure ofRGS12 PDZ domain | | 分子名称: | Regulator of G-protein signaling 12 | | 著者 | Lu, G, Ji, P, Huang, H. | | 登録日 | 2010-03-10 | | 公開日 | 2011-03-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Determining the Molecular Basis for the pH-dependent
Interaction between the PDZ of Human RGS12 and MEK2
To be Published
|
|
2GYT
 
 | |
2PPB
 
 | | Crystal structure of the T. thermophilus RNAP polymerase elongation complex with the ntp substrate analog and antibiotic streptolydigin | | 分子名称: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA (5'-D(*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*AP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*TP*GP*TP*CP*TP*GP*GP*CP*GP*TP*TP*CP*GP*CP*GP*CP*GP*CP*CP*G)-3'), ... | | 著者 | Vassylyev, D.G, Vassylyeva, M.N, Artsimovitch, I, Landick, R. | | 登録日 | 2007-04-28 | | 公開日 | 2007-07-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for substrate loading in bacterial RNA polymerase.
Nature, 448, 2007
|
|
5XGH
 
 | | Crystal structure of PI3K complex with an inhibitor | | 分子名称: | 3-[(4-fluorophenyl)methylamino]-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | 著者 | Song, K, Yang, X, Zhao, Y, Jian, Z. | | 登録日 | 2017-04-13 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | New Insights into PI3K Inhibitor Design using X-ray Structures of PI3K alpha Complexed with a Potent Lead Compound.
Sci Rep, 7, 2017
|
|
2LB5
 
 | | Refined Structural Basis for the Photoconversion of A Phytochrome to the Activated FAR-RED LIGHT-ABSORBING Form | | 分子名称: | PHYCOCYANOBILIN, Sensor histidine kinase | | 著者 | Cornilescu, C.C, Cornilescu, G, Ulijasz, A.T, Vierstra, R.D, Markley, J.L. | | 登録日 | 2011-03-23 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the photoconversion of a phytochrome to the activated Pfr form.
Nature, 463, 2010
|
|
2LHR
 
 | |
2MBK
 
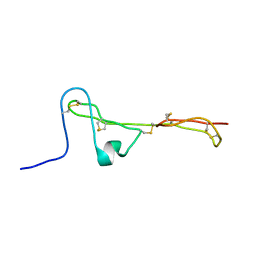 | | The Clip-segment of the von Willebrand domain 1 of the BMP modulator protein Crossveinless 2 is preformed | | 分子名称: | Crossveinless 2 | | 著者 | Mueller, T.D, Fiebig, J.E, Weidauer, S.E, Qiu, L, Bauer, M, Schmieder, P, Beerbaum, M, Zhang, J, Oschkinat, H, Sebald, W. | | 登録日 | 2013-08-02 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Clip-Segment of the von Willebrand Domain 1 of the BMP Modulator Protein Crossveinless 2 Is Preformed.
Molecules, 18, 2013
|
|
7Y3L
 
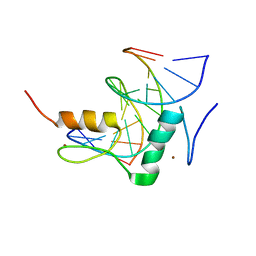 | | Structure of SALL3 ZFC4 bound with 12 bp AT-rich dsDNA | | 分子名称: | DNA (12-mer), Sal-like protein 3, ZINC ION | | 著者 | Ru, W, Xu, C. | | 登録日 | 2022-06-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3I
 
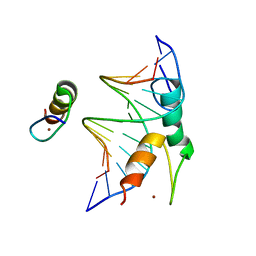 | | Structure of DNA bound SALL4 | | 分子名称: | DNA (12-mer), Sal-like protein 4, ZINC ION | | 著者 | Ru, W, Xu, C. | | 登録日 | 2022-06-10 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3K
 
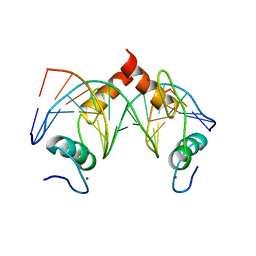 | | Structure of SALL4 ZFC4 bound with 16 bp AT-rich dsDNA | | 分子名称: | DNA (16-mer), Sal-like protein 4, ZINC ION | | 著者 | Ru, W, Xu, C. | | 登録日 | 2022-06-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3M
 
 | | Structure of SALL4 ZFC1 bound with 16 bp AT-rich dsDNA | | 分子名称: | DNA (16-mer), Sal-like protein 4, ZINC ION | | 著者 | Ru, W, Xu, C. | | 登録日 | 2022-06-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.723 Å) | | 主引用文献 | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7X78
 
 | | L-fuculose 1-phosphate aldolase | | 分子名称: | L-fuculose phosphate aldolase, MAGNESIUM ION, SULFATE ION | | 著者 | Lou, X, Zhang, Q, Bartlam, M. | | 登録日 | 2022-03-09 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural characterization of an L-fuculose-1-phosphate aldolase from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 607, 2022
|
|
6ISO
 
 | | Human SIRT3 Recognizing H3K4cr | | 分子名称: | (2E)-BUT-2-ENAL, ARG-THR-LYS-GLN-THR-ALA-ARG, GLYCEROL, ... | | 著者 | Wang, Y, Hao, Q. | | 登録日 | 2018-11-17 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach.
Elife, 3, 2014
|
|
4LCZ
 
 | | Crystal structure of a multilayer-packed major light-harvesting complex | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | Wan, T, Li, M, Chang, W.R. | | 登録日 | 2013-06-24 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of a multilayer packed major light-harvesting complex: implications for grana stacking in higher plants.
Mol Plant, 7, 2014
|
|
7EOQ
 
 | |
7EOT
 
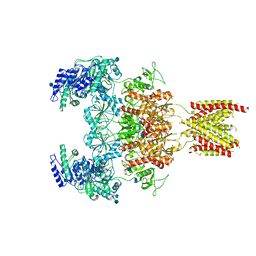 | |
7EOU
 
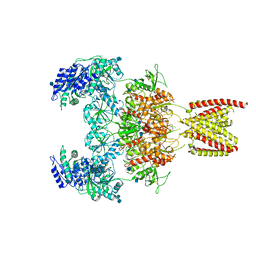 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901/9-AA bound state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, 9-AMINOACRIDINE, ... | | 著者 | Wang, H, Zhu, S. | | 登録日 | 2021-04-22 | | 公開日 | 2021-06-30 | | 最終更新日 | 2021-08-18 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
7EOS
 
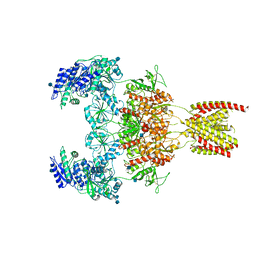 | |
7EOR
 
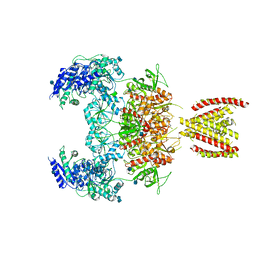 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901 bound state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, Glutamate receptor ionotropic, ... | | 著者 | Wang, H, Zhu, S. | | 登録日 | 2021-04-22 | | 公開日 | 2021-06-30 | | 最終更新日 | 2021-08-18 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
7X38
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 8A10 (CVB1-E:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3F
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 9A3 (CVB1-A:9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X42
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP0, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-01 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2I
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A particle in complex with nAb 2E6 (CVB1-pre-A:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-25 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2W
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A particle in complex with nAb 8A10 (CVB1-pre-A:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-26 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2T
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (CVB1-M:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-26 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.69 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
