7CTQ
 
 | | Peptidyl tryptophan dihydroxylase QhpG essential for tryptophylquinone cofactor biogenesis | | 分子名称: | (2~{R},3~{R},4~{S},5~{S},6~{R})-2-[(2~{R},3~{S},4~{R},5~{R},6~{R})-6-(cyclohexylmethoxy)-2-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-3-yl]oxy-6-(hydroxymethyl)oxane-3,4,5-triol, FLAVIN-ADENINE DINUCLEOTIDE, HEXANE-1,6-DIOL, ... | | 著者 | Oozeki, T, Nakai, T, Okajima, T. | | 登録日 | 2020-08-20 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.978 Å) | | 主引用文献 | Functional and structural characterization of a flavoprotein monooxygenase essential for biogenesis of tryptophylquinone cofactor.
Nat Commun, 12, 2021
|
|
5H35
 
 | | Crystal structures of the TRIC trimeric intracellular cation channel orthologue from Sulfolobus solfataricus | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Fab Heavy Chain, Fab Light Chain, ... | | 著者 | Kasuya, G, Hiraizumi, M, Hattori, M, Nureki, O. | | 登録日 | 2016-10-20 | | 公開日 | 2017-01-11 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (2.642 Å) | | 主引用文献 | Crystal structures of the TRIC trimeric intracellular cation channel orthologues
Cell Res., 26, 2016
|
|
7TBJ
 
 | | Composite structure of the human nuclear pore complex (NPC) symmetric core generated with a 12A cryo-ET map of the purified HeLa cell NPC | | 分子名称: | NUP107 CTD, NUP107 NTD, NUP133, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2022-06-29 | | 実験手法 | ELECTRON MICROSCOPY (23 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
1IDM
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE, LOOP-DELETED CHIMERA | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE | | 著者 | Sakurai, M, Ohzeki, M, Moriyama, H, Sato, M, Tanaka, N. | | 登録日 | 1995-05-19 | | 公開日 | 1995-09-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of a loop-deleted variant of 3-isopropylmalate dehydrogenase from Thermus thermophilus: an internal reprieve tolerance mechanism.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
5H2B
 
 | | Structure of a novel antibody G196 | | 分子名称: | G196 antibody Heavy chain, G196 antibody Light chain | | 著者 | Park, S.Y, Sugiyama, K. | | 登録日 | 2016-10-14 | | 公開日 | 2017-03-22 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | G196 epitope tag system: a novel monoclonal antibody, G196, recognizes the small, soluble peptide DLVPR with high affinity.
Sci Rep, 7, 2017
|
|
2CNP
 
 | |
3VVS
 
 | | Crystal structure of MATE in complex with MaD3S | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein, macrocyclic peptide | | 著者 | Tanaka, Y, Ishitani, R, Nureki, O. | | 登録日 | 2012-07-27 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3VVO
 
 | |
3VVN
 
 | |
3WBN
 
 | | Crystal structure of MATE in complex with MaL6 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MaL6, Putative uncharacterized protein | | 著者 | Tanaka, Y, Ishitani, R, Nureki, O. | | 登録日 | 2013-05-20 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3W4T
 
 | | Crystal structure of MATE P26A mutant | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein | | 著者 | Tanaka, Y, Ishitani, R, Nureki, O. | | 登録日 | 2013-01-16 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.096 Å) | | 主引用文献 | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
2LEX
 
 | | Complex of the C-terminal WRKY domain of AtWRKY4 and a W-box DNA | | 分子名称: | DNA (5'-D(*CP*G*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*C*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*CP*G)-3'), Probable WRKY transcription factor 4, ... | | 著者 | Yamasaki, K, Kigawa, T, Watanabe, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2011-06-24 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for sequence-spscific DNA recognition by an Arabidopsis WRKY transcription factor
J.Biol.Chem., 2012
|
|
6Z9C
 
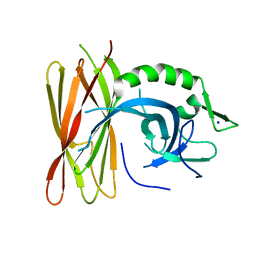 | | Structure of human POLDIP2, a multifaceted adaptor protein in metabolism and genome stability | | 分子名称: | Polymerase delta-interacting protein 2, SODIUM ION | | 著者 | Kulik, A.A, Maruszczak, K, Nabi, N.L.M, Bingham, R.J, Cooper, C.D.O. | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure and molecular dynamics of human POLDIP2, a multifaceted adaptor protein in metabolism and genome stability.
Protein Sci., 30, 2021
|
|
4NER
 
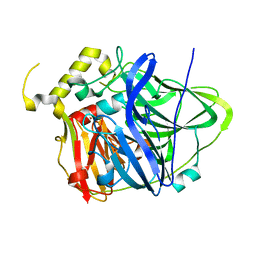 | | Multicopper Oxidase CueO (data1) | | 分子名称: | Blue copper oxidase CueO, COPPER (II) ION, HYDROXIDE ION, ... | | 著者 | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | 登録日 | 2013-10-30 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | New insights into the catalytic active-site structure of multicopper oxidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1WJ0
 
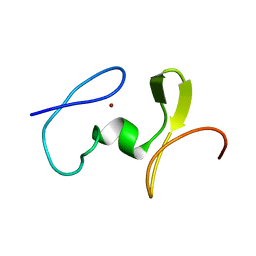 | |
1WIJ
 
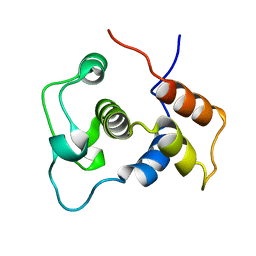 | | Solution Structure of the DNA-Binding Domain of Ethylene-Insensitive3-Like3 | | 分子名称: | ETHYLENE-INSENSITIVE3-like 3 protein | | 著者 | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-28 | | 公開日 | 2004-11-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3.
J.Mol.Biol., 348, 2005
|
|
1WJ2
 
 | | Solution Structure of the C-terminal WRKY Domain of AtWRKY4 | | 分子名称: | Probable WRKY transcription factor 4, ZINC ION | | 著者 | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-28 | | 公開日 | 2004-11-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an Arabidopsis WRKY DNA binding domain.
Plant Cell, 17, 2005
|
|
1WID
 
 | | Solution Structure of the B3 DNA-Binding Domain of RAV1 | | 分子名称: | DNA-binding protein RAV1 | | 著者 | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-28 | | 公開日 | 2004-11-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the B3 DNA Binding Domain of the Arabidopsis Cold-Responsive Transcription Factor RAV1
Plant Cell, 16, 2004
|
|
5BY6
 
 | | Crystal structure of Trichinella spiralis thymidylate synthase complexed with dUMP | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, ... | | 著者 | Dowiercial, A, Jarmula, A, Rypniewski, W, Fraczyk, T, Wilk, P, Rode, W. | | 登録日 | 2015-06-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of nematode (parasitic T. spiralis and free living C. elegans), compared to mammalian, thymidylate synthases (TS). Molecular docking and molecular dynamics simulations in search for nematode-specific inhibitors of TS.
J. Mol. Graph. Model., 77, 2017
|
|
1IPD
 
 | | THREE-DIMENSIONAL STRUCTURE OF A HIGHLY THERMOSTABLE ENZYME, 3-ISOPROPYLMALATE DEHYDROGENASE OF THERMUS THERMOPHILUS AT 2.2 ANGSTROMS RESOLUTION | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | 著者 | Imada, K, Sato, M, Tanaka, N, Katsube, Y, Matsuura, Y, Oshima, T. | | 登録日 | 1992-01-29 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution.
J.Mol.Biol., 222, 1991
|
|
1OSJ
 
 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE | | 著者 | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | 登録日 | 1996-10-22 | | 公開日 | 1997-01-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
1OSI
 
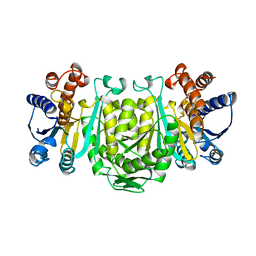 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE | | 著者 | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | 登録日 | 1996-10-22 | | 公開日 | 1997-01-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
1UL5
 
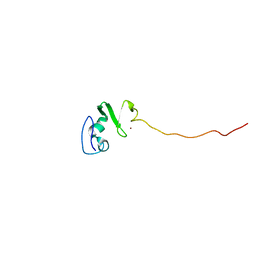 | | Solution structure of the DNA-binding domain of squamosa promoter binding protein-like 7 | | 分子名称: | ZINC ION, squamosa promoter binding protein-like 7 | | 著者 | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-09-09 | | 公開日 | 2004-03-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors.
J.Mol.Biol., 337, 2004
|
|
1UL4
 
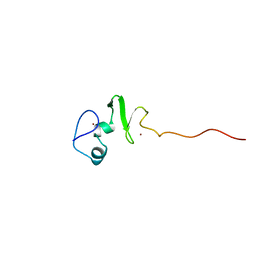 | | Solution structure of the DNA-binding domain of squamosa promoter binding protein-like 4 | | 分子名称: | ZINC ION, squamosa promoter binding protein-like 4 | | 著者 | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-09-09 | | 公開日 | 2004-03-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors.
J.Mol.Biol., 337, 2004
|
|
1XAB
 
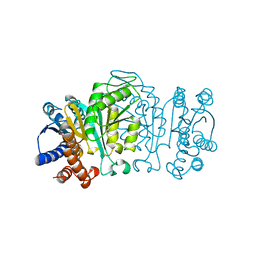 | |
