8GL3
 
 | |
3ET3
 
 | |
3ET0
 
 | |
3ET1
 
 | |
2F21
 
 | | human Pin1 Fip mutant | | 分子名称: | PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Jager, M, Zhang, Y, Bowman, M.E, Noel, J.P, Kelly, J.W. | | 登録日 | 2005-11-15 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6MHM
 
 | | Crystal structure of human acid ceramidase in covalent complex with carmofur | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Dementiev, A, Joachimiak, A, Doan, N. | | 登録日 | 2018-09-18 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.743 Å) | | 主引用文献 | Molecular Mechanism of Inhibition of Acid Ceramidase by Carmofur.
J. Med. Chem., 62, 2019
|
|
7Z2H
 
 | | Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine | | 分子名称: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, DNA (38-MER), Reverse transcriptase/ribonuclease H, ... | | 著者 | Singh, A.K, Das, K. | | 登録日 | 2022-02-27 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-10-12 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z2G
 
 | | Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine | | 分子名称: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, DNA (38-MER), Reverse transcriptase/ribonuclease H | | 著者 | Singh, A.K, Das, K. | | 登録日 | 2022-02-26 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-10-12 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z24
 
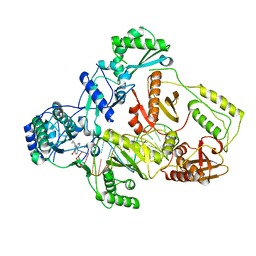 | | Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (38-mer), Reverse transcriptase/ribonuclease H | | 著者 | Singh, A.K, Das, K. | | 登録日 | 2022-02-25 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-03 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z2D
 
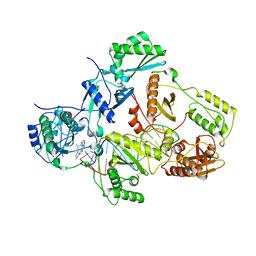 | |
7Z2E
 
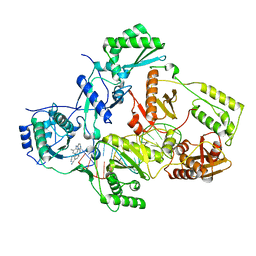 | |
7Z29
 
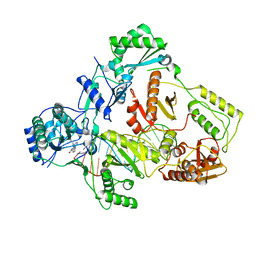 | | Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (38-MER), Reverse transcriptase/ribonuclease H, ... | | 著者 | Singh, A.K, Das, K. | | 登録日 | 2022-02-26 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-03 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3BEN
 
 | |
7KIS
 
 | | Crystal structure of Pseudomonas aeruginosa PBP2 in complex with WCK 5153 | | 分子名称: | (2S,5R)-1-formyl-N'-[(3R)-pyrrolidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, CHLORIDE ION, Peptidoglycan D,D-transpeptidase MrdA | | 著者 | Rajavel, M, van den Akker, F. | | 登録日 | 2020-10-24 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.869 Å) | | 主引用文献 | Structural Characterization of Diazabicyclooctane beta-Lactam "Enhancers" in Complex with Penicillin-Binding Proteins PBP2 and PBP3 of Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7KIV
 
 | |
7KIW
 
 | |
7KIT
 
 | |
