6EMF
 
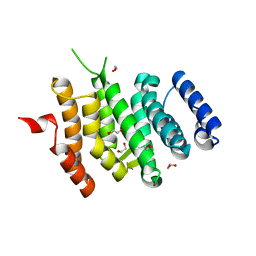 | |
6EMG
 
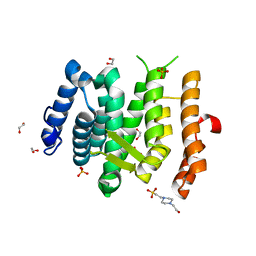 | |
4GMO
 
 | | Crystal structure of Syo1 | | 分子名称: | Putative uncharacterized protein | | 著者 | Bange, G, Sinning, I. | | 登録日 | 2012-08-16 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
4GMQ
 
 | | Ribosome-binding domain of Zuo1 | | 分子名称: | Putative ribosome associated protein | | 著者 | Kopp, J, Bange, G, Sinning, I. | | 登録日 | 2012-08-16 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural characterization of a eukaryotic chaperone-the ribosome-associated complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GMN
 
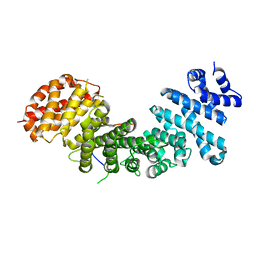 | |
4GNI
 
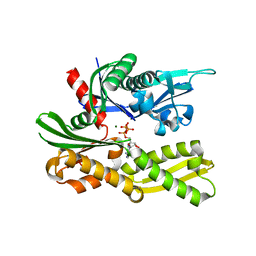 | |
4IPA
 
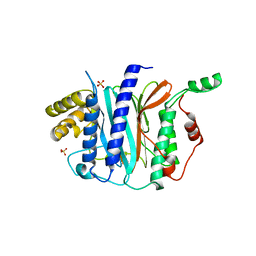 | | Structure of a thermophilic Arx1 | | 分子名称: | Putative curved DNA-binding protein, SULFATE ION | | 著者 | Bange, G, Sinning, I. | | 登録日 | 2013-01-09 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Consistent mutational paths predict eukaryotic thermostability.
BMC Evol Biol, 13, 2013
|
|
4P3F
 
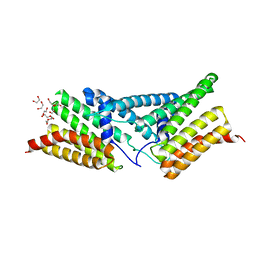 | | Structure of the human SRP68-RBD | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Signal recognition particle subunit SRP68 | | 著者 | Grotwinkel, J.T, Wild, K, Sinning, I. | | 登録日 | 2014-03-07 | | 公開日 | 2014-04-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.699 Å) | | 主引用文献 | SRP RNA remodeling by SRP68 explains its role in protein translocation.
Science, 344, 2014
|
|
4P3E
 
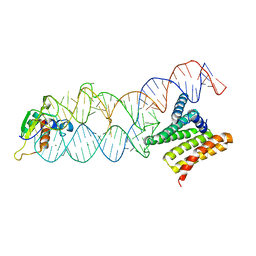 | | Structure of the human SRP S domain | | 分子名称: | MAGNESIUM ION, SRP RNA (124-mer), Signal recognition particle 19 kDa protein, ... | | 著者 | Grotwinkel, J.T, Wild, K, Sinning, I. | | 登録日 | 2014-03-07 | | 公開日 | 2014-04-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | SRP RNA remodeling by SRP68 explains its role in protein translocation.
Science, 344, 2014
|
|
4P3G
 
 | |
1JID
 
 | |
2YHS
 
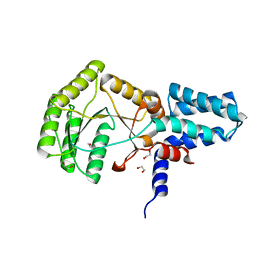 | | Structure of the E. coli SRP receptor FtsY | | 分子名称: | 1,2-ETHANEDIOL, CELL DIVISION PROTEIN FTSY | | 著者 | Stjepanovic, G, Bange, G, Wild, K, Sinning, I. | | 登録日 | 2011-05-05 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Lipids Trigger a Conformational Switch that Regulates Signal Recognition Particle (Srp)-Mediated Protein Targeting.
J.Biol.Chem., 286, 2011
|
|
3BS6
 
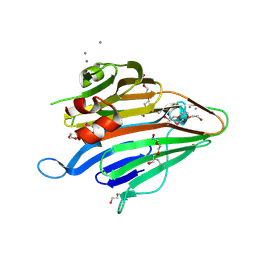 | |
5CK4
 
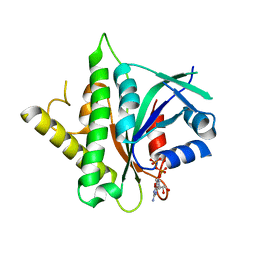 | |
5CK5
 
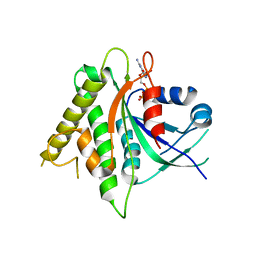 | |
5CK3
 
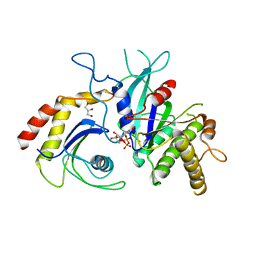 | | Signal recognition particle receptor SRb-GTP/SRX complex from Chaetomium thermophilum | | 分子名称: | GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Jadhav, B.R, Wild, K, Sinning, I. | | 登録日 | 2015-07-15 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure and Switch Cycle of SR beta as Ancestral Eukaryotic GTPase Associated with Secretory Membranes.
Structure, 23, 2015
|
|
5E4X
 
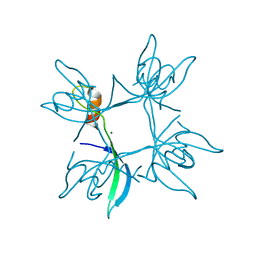 | | Crystal structure of cpSRP43 chromodomain 3 | | 分子名称: | MAGNESIUM ION, Signal recognition particle 43 kDa protein, chloroplastic | | 著者 | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | 登録日 | 2015-10-07 | | 公開日 | 2015-12-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
5E4W
 
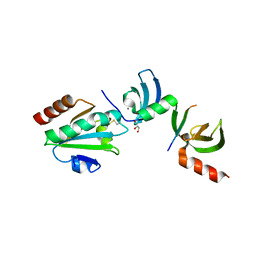 | | Crystal structure of cpSRP43 chromodomains 2 and 3 in complex with the Alb3 tail | | 分子名称: | CALCIUM ION, GLYCEROL, Inner membrane protein ALBINO3, ... | | 著者 | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | 登録日 | 2015-10-07 | | 公開日 | 2015-12-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
5EM2
 
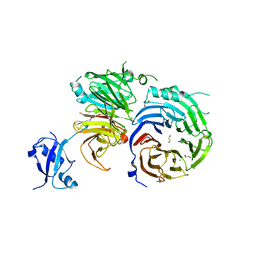 | | Crystal structure of the Erb1-Ytm1 complex | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ribosome biogenesis protein ERB1, ... | | 著者 | Ahmed, Y.L, Sinning, I. | | 登録日 | 2015-11-05 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface.
Nucleic Acids Res., 44, 2016
|
|
5BY8
 
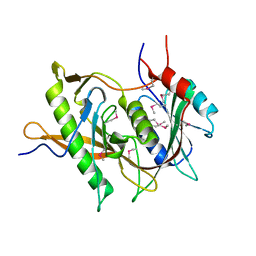 | | The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis | | 分子名称: | Rpf2, Rrs1 | | 著者 | Kharde, S, Calvino, F.R, Gumiero, A, Wild, K, Sinning, I. | | 登録日 | 2015-06-10 | | 公開日 | 2015-07-08 | | 最終更新日 | 2015-08-26 | | 実験手法 | X-RAY DIFFRACTION (1.515 Å) | | 主引用文献 | The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis.
Nucleic Acids Res., 43, 2015
|
|
2PX3
 
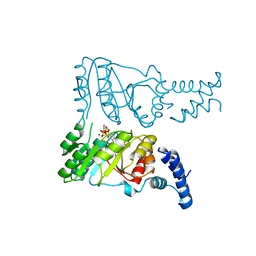 | |
2PX0
 
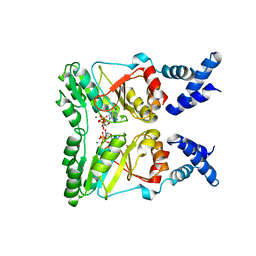 | | Crystal structure of FlhF complexed with GMPPNP/Mg(2+) | | 分子名称: | Flagellar biosynthesis protein flhF, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Bange, G, Wild, K, Sinning, I. | | 登録日 | 2007-05-14 | | 公開日 | 2007-09-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Q8K
 
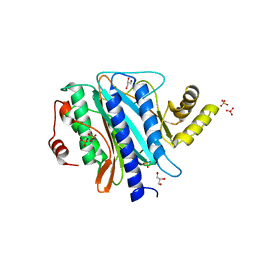 | | The crystal structure of Ebp1 | | 分子名称: | GLYCEROL, Proliferation-associated protein 2G4, SULFATE ION | | 著者 | Kowalinski, E, Bange, G, Wild, K, Sinning, I. | | 登録日 | 2007-06-11 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions.
Febs Lett., 581, 2007
|
|
6RY3
 
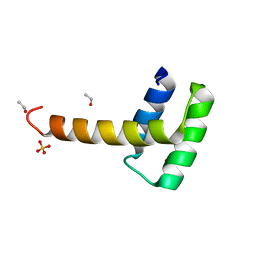 | | Structure of the WUS homeodomain | | 分子名称: | ACETYL GROUP, Protein WUSCHEL, SULFATE ION | | 著者 | Sloan, J.J, Wild, K, Sinning, I. | | 登録日 | 2019-06-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.374 Å) | | 主引用文献 | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
6SO5
 
 | | Homo sapiens WRB/CAML heterotetramer in complex with a TRC40 dimer | | 分子名称: | ATPase ASNA1, Calcium signal-modulating cyclophilin ligand, Tail-anchored protein insertion receptor WRB, ... | | 著者 | McDowell, M.A, Heimes, M, Wild, K, Flemming, D, Sinning, I. | | 登録日 | 2019-08-29 | | 公開日 | 2020-09-09 | | 最終更新日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural Basis of Tail-Anchored Membrane Protein Biogenesis by the GET Insertase Complex.
Mol.Cell, 80, 2020
|
|
