5LDT
 
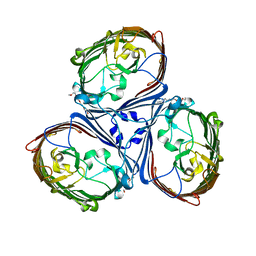 | | Crystal Structures of MOMP from Campylobacter jejuni | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, MOMP porin | | 著者 | Ferrara, L.G.M, Wallat, G.D, Moynie, L, Naismith, J.H. | | 登録日 | 2016-06-27 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
5LQ4
 
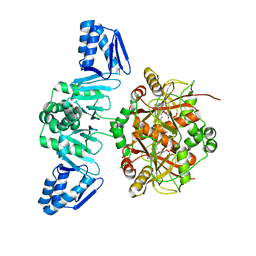 | | The Structure of ThcOx, the First Oxidase Protein from the Cyanobactin Pathways | | 分子名称: | CyaGox, FLAVIN MONONUCLEOTIDE | | 著者 | Bent, A.F, Wagner, A, Naismith, J.H. | | 登録日 | 2016-08-16 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure of the cyanobactin oxidase ThcOx from Cyanothece sp. PCC 7425, the first structure to be solved at Diamond Light Source beamline I23 by means of S-SAD.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5LMZ
 
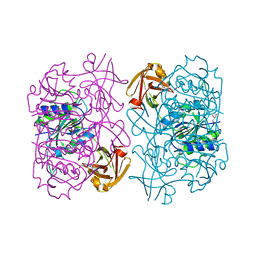 | | Fluorinase from Streptomyces sp. MA37 | | 分子名称: | 1-DEAZA-ADENOSINE, CHLORIDE ION, Fluorinase, ... | | 著者 | Mann, G, O'Hagan, D, Naismith, J.H, Bandaranayaka, N. | | 登録日 | 2016-08-02 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining.
Chembiochem, 15, 2014
|
|
5LDV
 
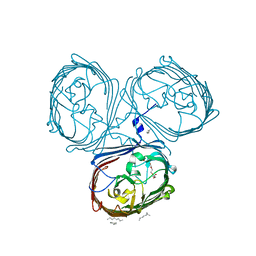 | | Crystal Structures of MOMP from Campylobacter jejuni | | 分子名称: | CALCIUM ION, MOMP porin, N-OCTANE, ... | | 著者 | Wallat, G.D, Ferrara, L.M.G, Moynie, L, Naismith, J.H. | | 登録日 | 2016-06-28 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
3LCC
 
 | |
3OHL
 
 | |
3OHO
 
 | |
1G0R
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). THYMIDINE/GLUCOSE-1-PHOSPHATE COMPLEX. | | 分子名称: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION, ... | | 著者 | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | 登録日 | 2000-10-07 | | 公開日 | 2000-12-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G1A
 
 | | THE CRYSTAL STRUCTURE OF DTDP-D-GLUCOSE 4,6-DEHYDRATASE (RMLB)FROM SALMONELLA ENTERICA SEROVAR TYPHIMURIUM | | 分子名称: | DTDP-D-GLUCOSE 4,6-DEHYDRATASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Allard, S.T.M, Giraud, M.-F, Whitfield, C, Graninger, M, Messner, P, Naismith, J.H. | | 登録日 | 2000-10-11 | | 公開日 | 2001-03-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | The crystal structure of dTDP-D-Glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP-l-rhamnose pathway.
J.Mol.Biol., 307, 2001
|
|
1G2V
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TTP COMPLEX. | | 分子名称: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, THYMIDINE-5'-TRIPHOSPHATE | | 著者 | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | 登録日 | 2000-10-21 | | 公開日 | 2000-12-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G3L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-L-RHAMNOSE COMPLEX. | | 分子名称: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | 著者 | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | 登録日 | 2000-10-24 | | 公開日 | 2000-12-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G23
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). GLUCOSE-1-PHOSPHATE COMPLEX. | | 分子名称: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | 著者 | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | 登録日 | 2000-10-16 | | 公開日 | 2000-12-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1FXO
 
 | |
1FZW
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). APO ENZYME. | | 分子名称: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | 著者 | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | 登録日 | 2000-10-04 | | 公開日 | 2000-12-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
5AIT
 
 | | A complex of of RNF4-RING domain, UbeV2, Ubc13-Ub (isopeptide crosslink) | | 分子名称: | E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, UBIQUITIN-CONJUGATING ENZYME E2 N, ... | | 著者 | Branigan, E, Naismith, J.H. | | 登録日 | 2015-02-17 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
5AIU
 
 | | A complex of RNF4-RING domain, Ubc13-Ub (isopeptide crosslink) | | 分子名称: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, ... | | 著者 | Branigan, E, Naismith, J.H. | | 登録日 | 2015-02-17 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
3TEK
 
 | | ThermoDBP: a non-canonical single-stranded DNA binding protein with a novel structure and mechanism | | 分子名称: | ThermoDBP-single stranded DNA binding protein | | 著者 | White, M.F, Paytubi, S, Liu, H, Graham, S, McMahon, S.A, Naismith, J.H. | | 登録日 | 2011-08-15 | | 公開日 | 2011-11-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Displacement of the canonical single-stranded DNA-binding protein in the Thermoproteales.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1SCR
 
 | | HIGH-RESOLUTION STRUCTURES OF SINGLE-METAL-SUBSTITUTED CONCANAVALIN A: THE CO,CA-PROTEIN AT 1.6 ANGSTROMS AND THE NI,CA-PROTEIN AT 2.0 ANGSTROMS | | 分子名称: | CALCIUM ION, CONCANAVALIN A, NICKEL (II) ION | | 著者 | Emmerich, C, Helliwell, J.R, Redshaw, M, Naismith, J.H, Harrop, S.J, Raftery, J, Kalb, A.J, Yariv, J, Dauter, Z, Wilson, K.S. | | 登録日 | 1993-12-06 | | 公開日 | 1994-05-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | High-resolution structures of single-metal-substituted concanavalin A: the Co,Ca-protein at 1.6 A and the Ni,Ca-protein at 2.0 A.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1SCS
 
 | | HIGH-RESOLUTION STRUCTURES OF SINGLE-METAL-SUBSTITUTED CONCANAVALIN A: THE CO,CA-PROTEIN AT 1.6 ANGSTROMS AND THE NI,CA-PROTEIN AT 2.0 ANGSTROMS | | 分子名称: | CALCIUM ION, COBALT (II) ION, CONCANAVALIN A | | 著者 | Emmerich, C, Helliwell, J.R, Redshaw, M, Naismith, J.H, Harrop, S.J, Raftery, J, Kalb, A.J, Yariv, J, Dauter, Z, Wilson, K.S. | | 登録日 | 1993-12-06 | | 公開日 | 1994-05-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | High-resolution structures of single-metal-substituted concanavalin A: the Co,Ca-protein at 1.6 A and the Ni,Ca-protein at 2.0 A.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1RQR
 
 | | Crystal structure and mechanism of a bacterial fluorinating enzyme, product complex | | 分子名称: | 5'-FLUORO-5'-DEOXYADENOSINE, 5'-fluoro-5'-deoxyadenosine synthase, METHIONINE | | 著者 | Dong, C, Huang, F, Deng, H, Schaffrath, C, Spencer, J.B, O'Hagan, D, Naismith, J.H. | | 登録日 | 2003-12-07 | | 公開日 | 2004-03-02 | | 最終更新日 | 2022-06-15 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Crystal structure and mechanism of a bacterial fluorinating enzyme
Nature, 427, 2004
|
|
1RQP
 
 | | Crystal structure and mechanism of a bacterial fluorinating enzyme | | 分子名称: | 5'-fluoro-5'-deoxyadenosine synthase, S-ADENOSYLMETHIONINE | | 著者 | Dong, C, Huang, F, Deng, H, Schaffrath, C, Spencer, J.B, O'Hagan, D, Naismith, J.H. | | 登録日 | 2003-12-06 | | 公開日 | 2004-03-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure and mechanism of a bacterial fluorinating enzyme
Nature, 427, 2004
|
|
1V0J
 
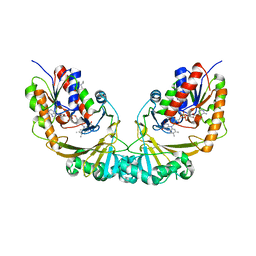 | | Udp-galactopyranose mutase from Mycobacterium tuberculosis | | 分子名称: | BICINE, FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | 著者 | Beis, K, Naismith, J.H. | | 登録日 | 2004-03-30 | | 公開日 | 2005-01-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidised state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state.
J. Mol. Biol., 348, 2005
|
|
8OWV
 
 | |
8OWW
 
 | |
8OWT
 
 | |
