4BK0
 
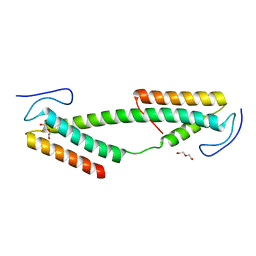 | | Crystal structure of the KIX domain of human RECQL5 (domain-swapped dimer) | | 分子名称: | ATP-DEPENDENT DNA HELICASE Q5, DI(HYDROXYETHYL)ETHER | | 著者 | Kassube, S.A, Jinek, M, Fang, J, Tsutakawa, S, Nogales, E. | | 登録日 | 2013-04-21 | | 公開日 | 2013-06-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Mimicry in Transcription Regulation of Human RNA Polymerase II by the DNA Helicase Recql5
Nat.Struct.Mol.Biol., 20, 2013
|
|
3P3F
 
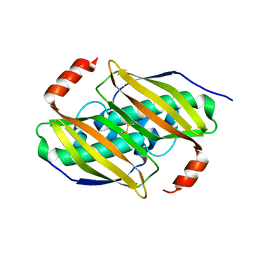 | | Crystal structure of the F36A mutant of the fluoroacetyl-CoA-specific thioesterase FlK | | 分子名称: | Fluoroacetyl coenzyme A thioesterase | | 著者 | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | 登録日 | 2010-10-04 | | 公開日 | 2010-10-20 | | 最終更新日 | 2011-11-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
3P2Q
 
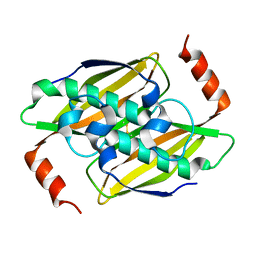 | | Crystal structure of the fluoroacetyl-CoA-specific thioesterase, FlK | | 分子名称: | Fluoroacetyl coenzyme A thioesterase | | 著者 | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | 登録日 | 2010-10-03 | | 公開日 | 2010-10-20 | | 最終更新日 | 2011-11-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
1R4P
 
 | | Shiga toxin type 2 | | 分子名称: | 1,2-ETHANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, FORMIC ACID, ... | | 著者 | Fraser, M.E, Fujinaga, M, Cherney, M.M, Melton-Celsa, A.R, Twiddy, E.M, O'Brien, A.D, James, M.N.G. | | 登録日 | 2003-10-07 | | 公開日 | 2004-05-11 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structure of Shiga Toxin Type 2 (Stx2) from Escherichia coli O157:H7.
J.Biol.Chem., 279, 2004
|
|
1R4Q
 
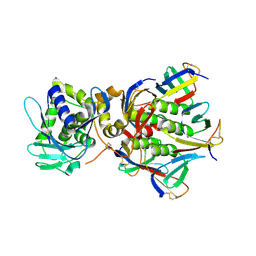 | | Shiga toxin | | 分子名称: | SHT cytotoxin A subunit, Shigella toxin chain B | | 著者 | Fraser, M.E, Fujinaga, M, Cherney, M.M, Melton-Celsa, A.R, Twiddy, E.M, O'Brien, A.D, James, M.N.G. | | 登録日 | 2003-10-07 | | 公開日 | 2004-05-11 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of Shiga Toxin Type 2 (Stx2) from Escherichia coli O157:H7.
J.Biol.Chem., 279, 2004
|
|
2XLJ
 
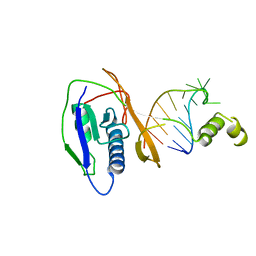 | | Crystal structure of the Csy4-crRNA complex, hexagonal form | | 分子名称: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | 著者 | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | 登録日 | 2010-07-20 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
2XLK
 
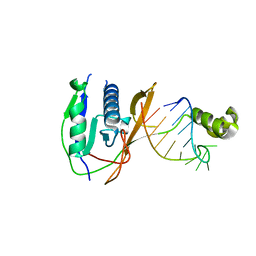 | | Crystal structure of the Csy4-crRNA complex, orthorhombic form | | 分子名称: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | 著者 | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | 登録日 | 2010-07-21 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.805 Å) | | 主引用文献 | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
2XLI
 
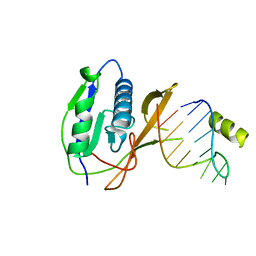 | | Crystal structure of the Csy4-crRNA complex, monoclinic form | | 分子名称: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | 著者 | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | 登録日 | 2010-07-20 | | 公開日 | 2010-09-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
2Y8Y
 
 | |
2Y8W
 
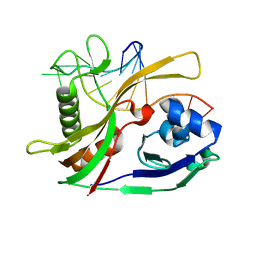 | |
2Y9H
 
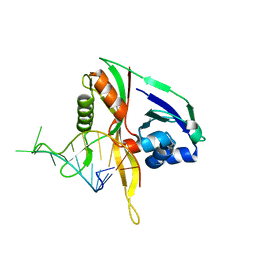 | |
7Z4E
 
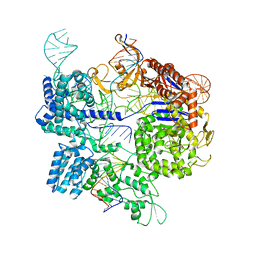 | | SpCas9 bound to 8-nucleotide complementary DNA substrate | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 8 nucleotide complementary DNA substrate, Target strand of 8 nucleotide complementary DNA substrate, ... | | 著者 | Pacesa, M, Jinek, M. | | 登録日 | 2022-03-03 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4I
 
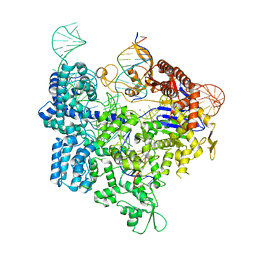 | |
7Z4C
 
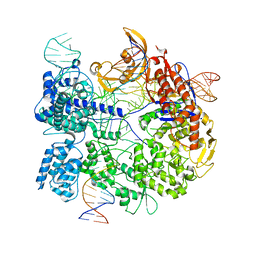 | | SpCas9 bound to 6 nucleotide complementary DNA substrate | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 6 nucleotide complementary DNA substrate, Target strand of 6 nucleotide complementary DNA substrate, ... | | 著者 | Pacesa, M, Jinek, M. | | 登録日 | 2022-03-03 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4G
 
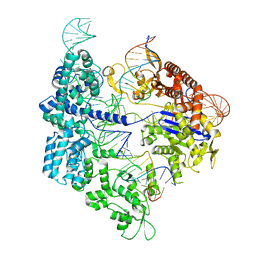 | | SpCas9 bound to 12-nucleotide complementary DNA substrate | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 12-nucleotide complementary DNA substrate, Target strand of 12-nucleotide complementary DNA substrate, ... | | 著者 | Pacesa, M, Jinek, M. | | 登録日 | 2022-03-03 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4K
 
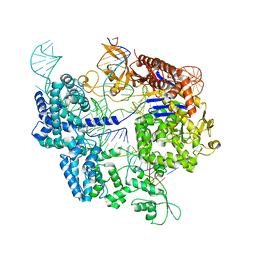 | | SpCas9 bound to 10-nucleotide complementary DNA substrate | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 10-nucleotide complementary DNA substrate, Target strand of 10-nucleotide complementary DNA substrate, ... | | 著者 | Pacesa, M, Jinek, M. | | 登録日 | 2022-03-04 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.81 Å) | | 主引用文献 | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4H
 
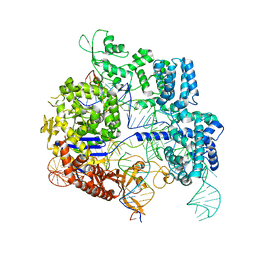 | | SpCas9 bound to 14-nucleotide complementary DNA substrate | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 14-nucleotide complementary DNA substrate, Target strand of 14-nucleotide complementary DNA substrate, ... | | 著者 | Pacesa, M, Jinek, M. | | 登録日 | 2022-03-03 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4J
 
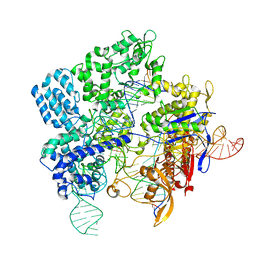 | |
7Z4L
 
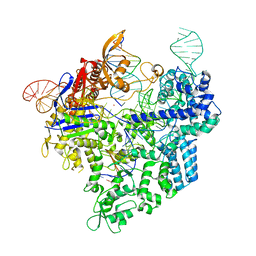 | |
5FSH
 
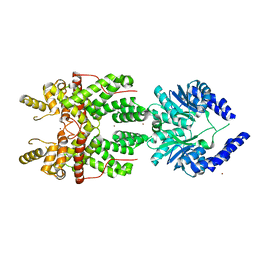 | |
5FW1
 
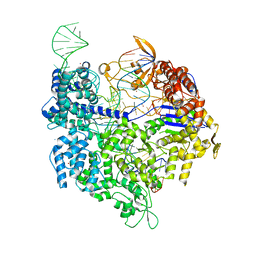 | | Crystal structure of SpyCas9 variant VQR bound to sgRNA and TGAG PAM target DNA | | 分子名称: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | 著者 | Anders, C, Bargsten, K, Jinek, M. | | 登録日 | 2016-02-11 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.499 Å) | | 主引用文献 | Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol.Cell, 61, 2016
|
|
5FW3
 
 | | Crystal structure of SpCas9 variant VRER bound to sgRNA and TGCG PAM target DNA | | 分子名称: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | 著者 | Anders, C, Bargsten, K, Jinek, M. | | 登録日 | 2016-02-11 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol.Cell, 61, 2016
|
|
5FW2
 
 | | Crystal structure of SpCas9 variant EQR bound to sgRNA and TGAG PAM target DNA | | 分子名称: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | 著者 | Anders, C, Bargsten, K, Jinek, M. | | 登録日 | 2016-02-11 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.676 Å) | | 主引用文献 | Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol.Cell, 61, 2016
|
|
6I0V
 
 | |
6I0S
 
 | | Crystal structure of DmTailor in complex with UMPNPP | | 分子名称: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, Terminal uridylyltransferase Tailor | | 著者 | Kroupova, A, Ivascu, A, Jinek, M. | | 登録日 | 2018-10-26 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
