7KLW
 
 | | Crystal structure of synthetic nanobody (Sb45+Sb68) complexes with SARS-CoV-2 receptor binding domain | | 分子名称: | SB45, Synthetic Nanobody, SB68, ... | | 著者 | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2020-11-01 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7MFU
 
 | | Crystal structure of synthetic nanobody (Sb14+Sb68) complexes with SARS-CoV-2 receptor binding domain | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Spike protein S1, ... | | 著者 | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2021-04-11 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7MFV
 
 | | Crystal structure of synthetic nanobody (Sb16) | | 分子名称: | 1,2-ETHANEDIOL, Synthetic Nanobody #16 (Sb16) | | 著者 | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2021-04-11 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
6M08
 
 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor. | | 分子名称: | (2S)-2-[(Z)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | 著者 | Hu, H.C, Xu, Y.C. | | 登録日 | 2020-02-20 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
6M06
 
 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | 分子名称: | (2S)-2-[(Z)-1,3-bis(oxidanyl)-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | 著者 | Hu, H.C, Xu, Y.C. | | 登録日 | 2020-02-20 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
6M07
 
 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | 分子名称: | (2S)-2-[(E)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)-2-[2,2,2-tris(fluoranyl)ethoxy]phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | 著者 | Hu, H.C, Xu, Y.C. | | 登録日 | 2020-02-20 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
7SSX
 
 | | Structure of human Kv1.3 | | 分子名称: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, Green fluorescent protein fusion | | 著者 | Meyerson, J.R, Selvakumar, P. | | 登録日 | 2021-11-11 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSZ
 
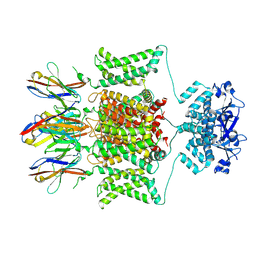 | |
7SSY
 
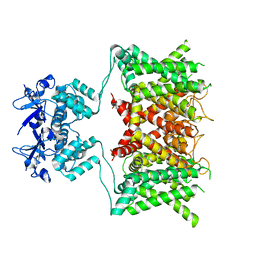 | |
5UBB
 
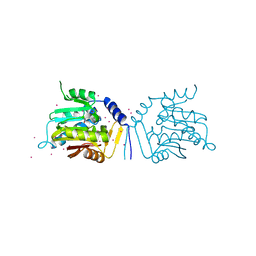 | | Crystal structure of human alpha N-terminal protein methyltransferase 1B | | 分子名称: | Alpha N-terminal protein methyltransferase 1B, S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION | | 著者 | Dong, C, Zhu, L, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2016-12-20 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | An asparagine/glycine switch governs product specificity of human N-terminal methyltransferase NTMT2.
Commun Biol, 1, 2018
|
|
6B6H
 
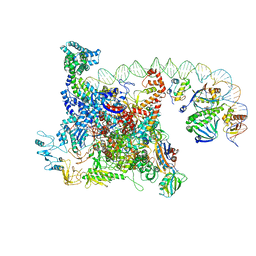 | | The cryo-EM structure of a bacterial class I transcription activation complex | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Liu, B, Hong, C, Huang, R, Yu, Z, Steitz, T.A. | | 登録日 | 2017-10-02 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis of bacterial transcription activation.
Science, 358, 2017
|
|
7VMB
 
 | |
5ZBE
 
 | | Crystal structure of AerE from Microcystis aeruginosa | | 分子名称: | Cupin domain protein, FE (II) ION, TRIETHYLENE GLYCOL | | 著者 | Qiu, X. | | 登録日 | 2018-02-11 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and functional insights into the role of a cupin superfamily isomerase in the biosynthesis of Choi moiety of aeruginosin.
J. Struct. Biol., 205, 2019
|
|
5ZBF
 
 | |
2OUL
 
 | | The Structure of Chagasin in Complex with a Cysteine Protease Clarifies the Binding Mode and Evolution of a New Inhibitor Family | | 分子名称: | Chagasin, Falcipain 2 | | 著者 | Wang, S.X, Chand, K, Huang, R, Whisstock, J, Jacobelli, J, Fletterick, R.J, Rosenthal, P.J, McKerrow, J.H. | | 登録日 | 2007-02-11 | | 公開日 | 2008-02-26 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The structure of chagasin in complex with a cysteine protease clarifies the binding mode and evolution of an inhibitor family.
Structure, 15, 2007
|
|
7KGK
 
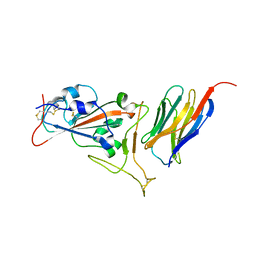 | | Crystal structure of synthetic nanobody (Sb16) complexes with SARS-CoV-2 receptor binding domain | | 分子名称: | Sb16, Sybody-16, Synthetic Nanobody, ... | | 著者 | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2020-10-16 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7TLJ
 
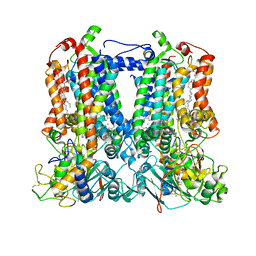 | | Rhodobacter sphaeroides Mitochondrial respiratory chain complex | | 分子名称: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, (5S)-3-anilino-5-methyl-5-(6-phenoxypyridin-3-yl)-1,3-oxazolidine-2,4-dione, 14 kDa peptide of ubiquinol-cytochrome c2 oxidoreductase complex, ... | | 著者 | Xia, D, Zhou, F, Esser, L, Huang, R. | | 登録日 | 2022-01-18 | | 公開日 | 2023-02-08 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Conformation Switch of Rieske ISP subunit is revealed by the Crystal Structure of Bacterial Cytochrome bc1 in Complex with Azoxystrobin
to be published
|
|
