6ZLL
 
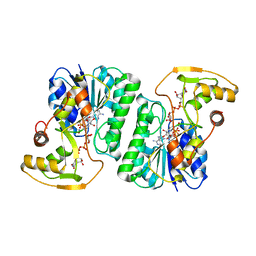 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Galacturonic acid and NAD | | 分子名称: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Iacovino, L.G, Savino, S, Mattevi, A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
6ZL6
 
 | |
6ZLA
 
 | |
6ZLK
 
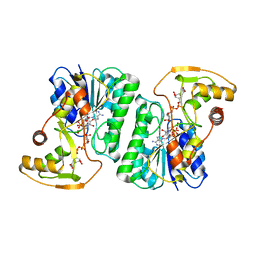 | | Equilibrium Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Glucuronic acid/UDP-Galacturonic acid and NAD | | 分子名称: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Iacovino, L.G, Mattevi, A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
6TQ8
 
 | |
6TQ5
 
 | | Alcohol dehydrogenase from Candida magnoliae DSMZ 70638 (ADHA): complex with NADP+ | | 分子名称: | Enzyme subunit, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Rovida, S, Aalbers, F.S, Fraaije, M.W, Mattevi, A. | | 登録日 | 2019-12-16 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Approaching boiling point stability of an alcohol dehydrogenase through computationally-guided enzyme engineering.
Elife, 9, 2020
|
|
6TQ3
 
 | |
5L3E
 
 | | LSD1-CoREST1 in complex with quinazoline-derivative reversible inhibitor | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6,7-dimethoxyquinazoline-2,4-diamine, ... | | 著者 | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | 登録日 | 2016-04-10 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
5L3F
 
 | | LSD1-CoREST1 in complex with polymyxin B | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, Polmyxin B, ... | | 著者 | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | 登録日 | 2016-04-10 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
5L3G
 
 | | LSD1-CoREST1 in complex with polymyxin E (colistin) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | 著者 | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | 登録日 | 2016-04-10 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
5LBQ
 
 | | LSD1-CoREST1 in complex with quinazoline-derivative reversible inhibitor | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, N2-(3-(dimethylamino)propyl)-6,7-dimethoxy-N4,N4-dimethylquinazoline-2,4-diamine, ... | | 著者 | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | 登録日 | 2016-06-16 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
1RZ0
 
 | | Flavin reductase PheA2 in native state | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, phenol 2-hydroxylase component B | | 著者 | van den Heuvel, R.H, Westphal, A.H, Heck, A.J, Walsh, M.A, Rovida, S, van Berkel, W.J, Mattevi, A. | | 登録日 | 2003-12-23 | | 公開日 | 2004-04-06 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Studies on Flavin Reductase PheA2 Reveal Binding of NAD in an Unusual Folded Conformation and Support Novel Mechanism of Action.
J.Biol.Chem., 279, 2004
|
|
1RZ1
 
 | | Reduced flavin reductase PheA2 in complex with NAD | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, phenol 2-hydroxylase component B | | 著者 | Van Den Heuvel, R.H, Westphal, A.H, Heck, A.J, Walsh, M.A, Rovida, S, Van Berkel, W.J, Mattevi, A. | | 登録日 | 2003-12-23 | | 公開日 | 2004-04-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Studies on Flavin Reductase PheA2 Reveal Binding of NAD in an Unusual Folded Conformation and Support Novel Mechanism of Action.
J.Biol.Chem., 279, 2004
|
|
2C7G
 
 | | FprA from Mycobacterium tuberculosis: His57Gln mutant | | 分子名称: | 4-OXO-NICOTINAMIDE-ADENINE DINUCLEOTIDE PHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-FERREDOXIN REDUCTASE FPRA, ... | | 著者 | Pennati, A, Razeto, A, De Rosa, M, Pandini, V, Vanoni, M.A, Aliverti, A, Mattevi, A, Coda, A, Zanetti, G. | | 登録日 | 2005-11-24 | | 公開日 | 2006-07-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of the His57-Glu214 Ionic Couple Located in the Active Site of Mycobacterium Tuberculosis Fpra.
Biochemistry, 45, 2006
|
|
1KNP
 
 | |
1KNR
 
 | | L-aspartate oxidase: R386L mutant | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, L-aspartate oxidase, ... | | 著者 | Bossi, R.T, Mattevi, A. | | 登録日 | 2001-12-19 | | 公開日 | 2002-04-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of FAD-bound L-aspartate oxidase: insight into substrate specificity and catalysis.
Biochemistry, 41, 2002
|
|
6H0N
 
 | |
6HQG
 
 | |
6H0P
 
 | |
2XFU
 
 | | Human monoamine oxidase B with tranylcypromine | | 分子名称: | 3-PHENYLPROPANAL, Amine oxidase [flavin-containing] B, [[(2R,3S,4S)-5-[(4AS)-7,8-DIMETHYL-2,4-DIOXO-4A,5-DIHYDROBENZO[G]PTERIDIN-10-YL]-2,3,4-TRIHYDROXY-PENTOXY]-HYDROXY-PHOSPHORYL] [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL HYDROGEN PHOSPHATE | | 著者 | Binda, C, Li, M, Hubalek, F, Restelli, N, Edmondson, D.E, Mattevi, A. | | 登録日 | 2010-05-26 | | 公開日 | 2010-06-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Potentiation of ligand binding through cooperative effects in monoamine oxidase B.
J. Biol. Chem., 285, 2010
|
|
6HQD
 
 | |
6HQW
 
 | |
6HHE
 
 | | Crystal structure of the medfly Odorant Binding Protein CcapOBP22/CcapOBP69a | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, Odorant binding protein OBP69a, SULFATE ION | | 著者 | Falchetto, M, Ciossani, G, Nenci, S, Mattevi, A, Gasperi, G, Forneris, F. | | 登録日 | 2018-08-28 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.516 Å) | | 主引用文献 | Structural and biochemical evaluation of Ceratitis capitata odorant-binding protein 22 affinity for odorants involved in intersex communication.
Insect Mol.Biol., 28, 2019
|
|
5MLN
 
 | |
5MXU
 
 | | Structure of the Y503F mutant of vanillyl alcohol oxidase | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Vanillyl-alcohol oxidase | | 著者 | Ewing, T.A, Nguyen, Q.-T, Allan, R.C, Gygli, G, Romero, E, Binda, C, Fraaije, M.W, Mattevi, A, van Berkel, W.J.H. | | 登録日 | 2017-01-24 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Two tyrosine residues, Tyr-108 and Tyr-503, are responsible for the deprotonation of phenolic substrates in vanillyl-alcohol oxidase.
J. Biol. Chem., 292, 2017
|
|
