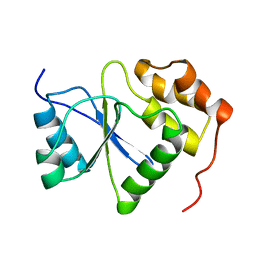
1V3A
 
 | |
4O23
 
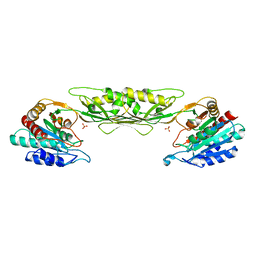 | | Crystal structure of mono-zinc form of succinyl diaminopimelate desuccinylase from Neisseria meningitidis MC58 | | 分子名称: | SULFATE ION, Succinyl-diaminopimelate desuccinylase, ZINC ION | | 著者 | Nocek, B, Holz, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-12-16 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Inhibition of the dapE-Encoded N-Succinyl-L,L-diaminopimelic Acid Desuccinylase from Neisseria meningitidis by L-Captopril.
Biochemistry, 54, 2015
|
|
4PPZ
 
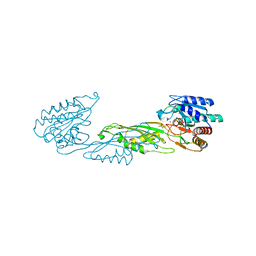 | | Crystal structure of zinc-bound succinyl-diaminopimelate desuccinylase from Neisseria meningitidis MC58 | | 分子名称: | PHOSPHATE ION, Succinyl-diaminopimelate desuccinylase, ZINC ION | | 著者 | Nocek, B, Holz, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-02-27 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Inhibition of the dapE-Encoded N-Succinyl-L,L-diaminopimelic Acid Desuccinylase from Neisseria meningitidis by L-Captopril.
Biochemistry, 54, 2015
|
|
4PQA
 
 | | Crystal Structure of succinyl-diaminopimelate desuccinylase from Neisseria meningitidis MC58 in complex with the Inhibitor Captopril | | 分子名称: | L-CAPTOPRIL, SULFATE ION, Succinyl-diaminopimelate desuccinylase, ... | | 著者 | Nocek, B, Starus, A, Holz, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-03-01 | | 公開日 | 2014-04-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Inhibition of the dapE-Encoded N-Succinyl-L,L-diaminopimelic Acid Desuccinylase from Neisseria meningitidis by L-Captopril.
Biochemistry, 54, 2015
|
|
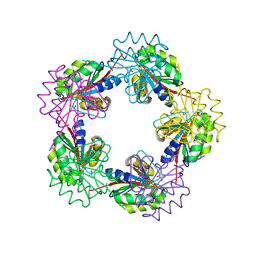
1OGF
 
 | |
1OGE
 
 | |
1OGC
 
 | |
1OGD
 
 | |
2PLN
 
 | |
1W01
 
 | |
1W00
 
 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | 分子名称: | STEROID DELTA-ISOMERASE | | 著者 | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | 登録日 | 2004-05-30 | | 公開日 | 2005-05-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
1W02
 
 | |
2P3U
 
 | |
1QJG
 
 | |
2P3T
 
 | |
4ZY4
 
 | |
4ZY6
 
 | |
1AYM
 
 | |
4ZY5
 
 | |
5BMS
 
 | |
8E1O
 
 | | Crystal structure of hTEAD2 bound to a methoxypyridine lipid pocket binder | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methoxy-N-({3-[2-(methylamino)-2-oxoethyl]phenyl}methyl)-4-{(E)-2-[trans-4-(trifluoromethyl)cyclohexyl]ethenyl}pyridine-2-carboxamide, Transcriptional enhancer factor TEF-4 | | 著者 | Noland, C.L, Dey, A, Zbieg, J, Crawford, J. | | 登録日 | 2022-08-10 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Targeting the Hippo pathway in cancers via ubiquitination dependent TEAD degradation
Biorxiv, 2024
|
|
4MLE
 
 | |
4MLH
 
 | |
5IME
 
 | | Crystal structure of P21-activated kinase 1 (PAK1) in complex with compound 9 | | 分子名称: | 8-(3-aminopropyl)-6-[2-chloro-4-(3-methyl-2-oxopyrazin-1(2H)-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase PAK 1 | | 著者 | Li, D, Wang, W. | | 登録日 | 2016-03-06 | | 公開日 | 2016-05-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.217 Å) | | 主引用文献 | Chemically Diverse Group I p21-Activated Kinase (PAK) Inhibitors Impart Acute Cardiovascular Toxicity with a Narrow Therapeutic Window.
J.Med.Chem., 59, 2016
|
|
7TYQ
 
 | | TEAD2 bound to Compound 1 | | 分子名称: | Transcriptional enhancer factor TEF-4, ethyl (8S)-7-oxo-5-[4-(trifluoromethyl)phenyl]-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carboxylate | | 著者 | Noland, C.L, Fong, R. | | 登録日 | 2022-02-14 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
