3FH0
 
 | | Crystal structure of putative universal stress protein KPN_01444 - ATPase | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, putative universal stress protein KPN_01444 | | 著者 | Chang, C, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-12-08 | | 公開日 | 2008-12-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of putative universal stress protein KPN_01444 - ATPase
To be Published
|
|
3F67
 
 | | Crystal Structure of Putative Dienelactone Hydrolase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, FORMIC ACID, ... | | 著者 | Kim, Y, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-11-05 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Crystal Structure of Putative Dienelactone Hydrolase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To be Published
|
|
3GL1
 
 | | Crystal structure of ATPase domain of Ssb1 chaperone, a member of the HSP70 family, from Saccharomyces cerevisiae | | 分子名称: | CHLORIDE ION, GLYCEROL, Heat shock protein SSB1, ... | | 著者 | Osipiuk, J, Li, H, Bargassa, M, Sahi, C, Craig, E.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-03-11 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Crystal structure of ATPase domain of Ssb1 chaperone, member of the HSP70 family from Saccharomyces cerevisiae.
To be Published
|
|
3FH2
 
 | |
3HJF
 
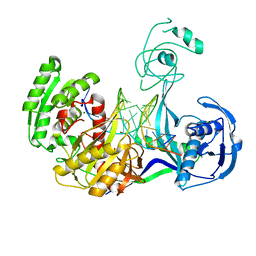 | | Crystal structure of T. thermophilus Argonaute E546 mutant protein complexed with DNA guide strand and 15-nt RNA target strand | | 分子名称: | 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', 5'-R(*CP*AP*AP*CP*CP*UP*AP*CP*UP*AP*CP*CP*UP*CP*G)-3', Argonaute, ... | | 著者 | Wang, Y, Li, H, Sheng, G, Patel, D.J. | | 登録日 | 2009-05-21 | | 公開日 | 2009-10-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.056 Å) | | 主引用文献 | Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes.
Nature, 461, 2009
|
|
3HJY
 
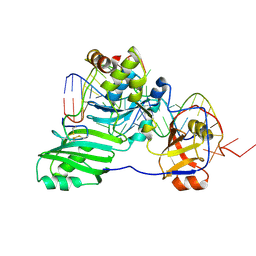 | | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA | | 分子名称: | 5'-R(*GP*GP*AP*GP*CP*GP*UP*GP*CP*GP*GP*UP*UP*U)-3', 5'-R(*GP*GP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*CP*CP*GP*CP*GP*GP*CP*GP*C)-3', RNA (25-MER), ... | | 著者 | Liang, B, Zhou, J, Kahen, E, Terns, R.M, Terns, M.P, Li, H. | | 登録日 | 2009-05-22 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HJW
 
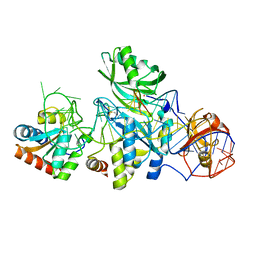 | | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA | | 分子名称: | 5'-R(*GP*AP*GP*CP*GP*(FHU)P*GP*CP*GP*GP*UP*UP*U)-3', 50S ribosomal protein L7Ae, POTASSIUM ION, ... | | 著者 | Liang, B, Zhou, J, Kahen, E, Terns, R.M, Terns, M.P, Li, H. | | 登録日 | 2009-05-22 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HK2
 
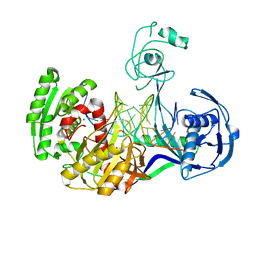 | | Crystal structure of T. thermophilus Argonaute N478 mutant protein complexed with DNA guide strand and 19-nt RNA target strand | | 分子名称: | 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', 5'-R(*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*AP*CP*CP*UP*CP*G)-3', Argonaute, ... | | 著者 | Wang, Y, Li, H, Sheng, G, Patel, D.J. | | 登録日 | 2009-05-22 | | 公開日 | 2009-10-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes.
Nature, 461, 2009
|
|
3IX7
 
 | |
3IF0
 
 | |
7KY7
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the apo E1 state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, CHOLESTEROL, ... | | 著者 | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | 登録日 | 2020-12-07 | | 公開日 | 2021-01-06 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
5BU9
 
 | | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 | | 分子名称: | Beta-N-acetylhexosaminidase, GLYCEROL | | 著者 | Chang, C, Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-06-03 | | 公開日 | 2015-06-17 | | 実験手法 | X-RAY DIFFRACTION (2.255 Å) | | 主引用文献 | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333
To Be Published
|
|
7KY6
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf1-Lem3 complex in the apo E1 state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | 著者 | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | 登録日 | 2020-12-07 | | 公開日 | 2021-01-06 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7KOE
 
 | | Electron bifurcating flavoprotein Fix/EtfABCX | | 分子名称: | Electron transfer flavoprotein, alpha subunit, beta subunit, ... | | 著者 | Feng, X, Li, H. | | 登録日 | 2020-11-08 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryoelectron microscopy structure and mechanism of the membrane-associated electron-bifurcating flavoprotein Fix/EtfABCX.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KYA
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the E2P state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | 著者 | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | 登録日 | 2020-12-07 | | 公開日 | 2021-01-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7KY8
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the E1-ATP state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | 著者 | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | 登録日 | 2020-12-07 | | 公開日 | 2021-01-06 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7KYC
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf1-Lem3 complex in the E2P state | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | 登録日 | 2020-12-07 | | 公開日 | 2021-01-06 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
3ILK
 
 | | The structure of a probable methylase family protein from Haemophilus influenzae Rd KW20 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | 著者 | Tan, K, Li, H, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-08-07 | | 公開日 | 2009-09-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | The structure of a probable methylase family protein from Haemophilus influenzae Rd KW20
To be Published
|
|
7KY5
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the E2P transition state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, CHOLESTEROL, ... | | 著者 | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | 登録日 | 2020-12-07 | | 公開日 | 2021-01-06 | | 実験手法 | ELECTRON MICROSCOPY (3.98 Å) | | 主引用文献 | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7KY9
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the E1-ADP state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | 登録日 | 2020-12-07 | | 公開日 | 2021-01-06 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7KYB
 
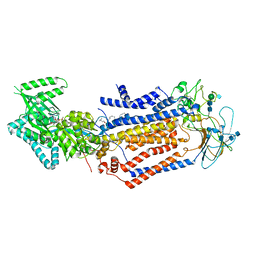 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf1-Lem3 complex in the E1-ADP state | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | 登録日 | 2020-12-07 | | 公開日 | 2021-01-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
3KBQ
 
 | |
5C11
 
 | |
5CD2
 
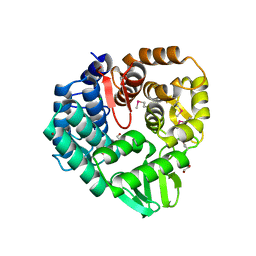 | | The crystal structure of endo-1,4-D-glucanase from Vibrio fischeri ES114 | | 分子名称: | CHLORIDE ION, Endo-1,4-D-glucanase, GLYCEROL, ... | | 著者 | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-07-02 | | 公開日 | 2015-07-22 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The crystal structure of endo-1,4-D-glucanase from Vibrio fischeri ES114
To Be Published
|
|
7KC0
 
 | | Structure of the Saccharomyces cerevisiae replicative polymerase delta in complex with a primer/template and the PCNA clamp | | 分子名称: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA (25-MER), DNA (5'-D(P*AP*TP*GP*AP*CP*CP*AP*TP*GP*AP*TP*TP*AP*CP*GP*AP*AP*TP*TP*GP*C)-3'), ... | | 著者 | Zheng, F, Georgescu, R, Li, H, O'Donnell, M.E. | | 登録日 | 2020-10-04 | | 公開日 | 2020-12-02 | | 最終更新日 | 2020-12-16 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of eukaryotic DNA polymerase delta bound to the PCNA clamp while encircling DNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
