7W6L
 
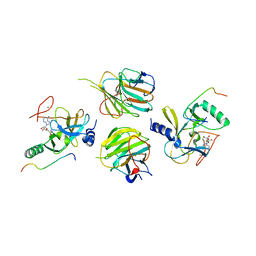 | | The crystal structure of MLL3-RBBP5-ASH2L in complex with H3K4me0 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6A
 
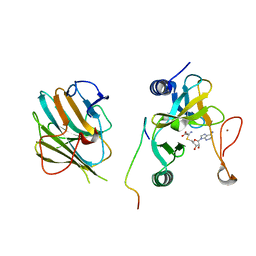 | | Crystal structure of the MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L complex | | 分子名称: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W67
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me0 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.194 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6J
 
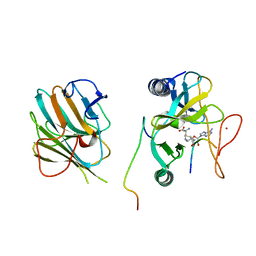 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me2 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7Y7M
 
 | |
7YBF
 
 | |
7YMS
 
 | |
7YRH
 
 | |
7YRF
 
 | |
7YV7
 
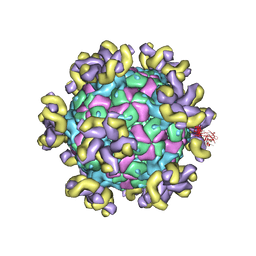 | |
7YV2
 
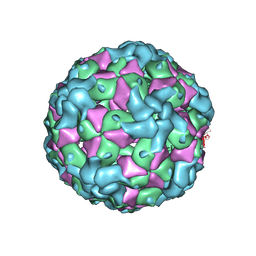 | |
2P6W
 
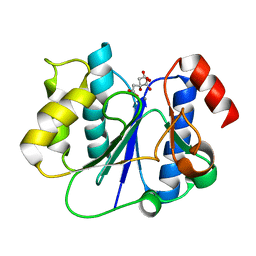 | | Crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | 分子名称: | CITRATE ANION, MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein | | 著者 | Zhang, Y, Ye, X, Van Etten, J.L, Rossmann, M.G. | | 登録日 | 2007-03-19 | | 公開日 | 2007-08-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
7V4L
 
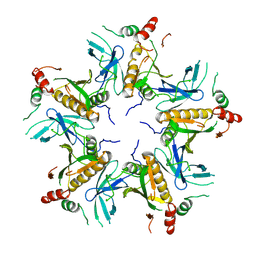 | |
7V4H
 
 | |
7V4J
 
 | |
7V4I
 
 | |
7V4K
 
 | |
