6AKG
 
 | |
6AKE
 
 | |
5WZR
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - Gal-NHAc-DNJ complex | | 分子名称: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, N-[(3S,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]acetamide, ... | | 著者 | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | 登録日 | 2017-01-18 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
6UT3
 
 | |
2LUE
 
 | | LC3B OPTN-LIR Ptot complex structure | | 分子名称: | Microtubule-associated proteins 1A/1B light chain 3B, Optineurin | | 著者 | Rogov, V.V, Rozenknop, A, Loehr, F, Guentert, P, Doetsch, V. | | 登録日 | 2012-06-13 | | 公開日 | 2013-07-17 | | 最終更新日 | 2022-08-24 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella.
Biochem.J., 454, 2013
|
|
6N2A
 
 | |
6N2F
 
 | |
1WWQ
 
 | | Solution Structure of Mouse ER | | 分子名称: | Enhancer of rudimentary homolog | | 著者 | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-01-12 | | 公開日 | 2006-01-03 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the mouse enhancer of rudimentary protein reveals a novel fold
J.Biomol.Nmr, 32, 2005
|
|
5B4O
 
 | | Crystal structure of Macrophage Migration Inhibitory Factor in complex with BTZO-14 | | 分子名称: | 1,2-ETHANEDIOL, 2-pyridin-3-yl-1,3-benzothiazin-4-one, Macrophage migration inhibitory factor, ... | | 著者 | Oki, H, Igaki, S, Moriya, Y, Hayano, Y, Habuka, N. | | 登録日 | 2016-04-07 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | BTZO-1, a cardioprotective agent, reveals that macrophage migration inhibitory factor regulates ARE-mediated gene expression
Chem. Biol., 17, 2010
|
|
5WZN
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - GalNAc complex | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Alpha-N-acetylgalactosaminidase, CALCIUM ION, ... | | 著者 | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | 登録日 | 2017-01-18 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZP
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - ligand free | | 分子名称: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, ZINC ION | | 著者 | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | 登録日 | 2017-01-18 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZQ
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - quadruple mutant | | 分子名称: | Alpha-N-acetylgalactosaminidase, GLYCEROL, ZINC ION | | 著者 | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | 登録日 | 2017-01-18 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
4YVF
 
 | | Structure of S-adenosyl-L-homocysteine hydrolase | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{[5-chloro-2-(4-chlorophenoxy)phenyl](2-{[2-(methylamino)ethyl]amino}-2-oxoethyl)amino}-N-(1,3-dihydro-2H-isoindol-2-yl)-N-methylacetamide, Adenosylhomocysteinase | | 著者 | Akiko, K. | | 登録日 | 2015-03-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery and structural analyses of S-adenosyl-L-homocysteine hydrolase inhibitors based on non-adenosine analogs.
Bioorg.Med.Chem., 23, 2015
|
|
5XDE
 
 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and dibenzothiophene | | 分子名称: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | 著者 | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | 登録日 | 2017-03-28 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
5XDG
 
 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and dibenzothiophene sulfoxide | | 分子名称: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | 著者 | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | 登録日 | 2017-03-28 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.747 Å) | | 主引用文献 | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
2QMC
 
 | |
2QM6
 
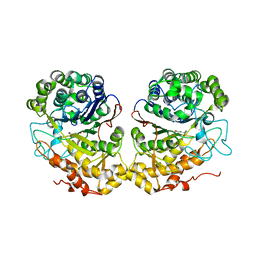 | |
5YLV
 
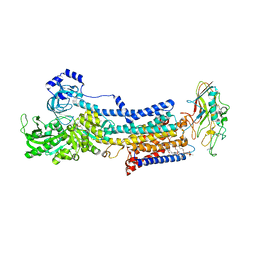 | | Crystal structure of the gastric proton pump complexed with SCH28080 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(2-methyl-8-phenylmethoxy-imidazo[1,2-a]pyridin-3-yl)ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Abe, K, Irie, K, Nakanishi, H, Fujiyoshi, Y. | | 登録日 | 2017-10-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.79977775 Å) | | 主引用文献 | Crystal structures of the gastric proton pump
Nature, 556, 2018
|
|
5YLU
 
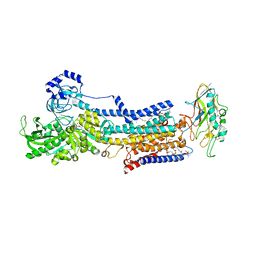 | | Crystal structure of the gastric proton pump complexed with vonoprazan | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[5-(2-fluorophenyl)-1-pyridin-3-ylsulfonyl-pyrrol-3-yl]-~{N}-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Abe, K, Irie, K, Nakanishi, H, Fujiyoshi, Y. | | 登録日 | 2017-10-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.79988956 Å) | | 主引用文献 | Crystal structures of the gastric proton pump
Nature, 556, 2018
|
|
6AKF
 
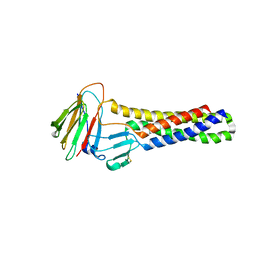 | |
2E0Y
 
 | |
2E0X
 
 | |
2E0W
 
 | |
5B5T
 
 | |
2E1T
 
 | | Crystal structure of Dendranthema morifolium DmAT complexed with malonyl-CoA | | 分子名称: | MALONYL-COENZYME A, acyl transferase | | 著者 | Unno, H, Ichimaida, F, Kusunoki, M, Nakayama, T. | | 登録日 | 2006-10-28 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and Mutational Studies of Anthocyanin Malonyltransferases Establish the Features of BAHD Enzyme Catalysis
J.Biol.Chem., 282, 2007
|
|
