7NAD
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Spb4 local refinement model | | 分子名称: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L17-A, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NAC
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Composite model | | 分子名称: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XKY
 
 | |
5DNI
 
 | | Crystal structure of Methanocaldococcus jannaschii Fumarate hydratase beta subunit | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Jayaraman, V, Kunala, J, Balaram, H. | | 登録日 | 2015-09-10 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Revisiting the Burden Borne by Fumarase: Enzymatic Hydration of an Olefin.
Biochemistry, 62, 2023
|
|
7D40
 
 | |
7D96
 
 | |
7D97
 
 | |
7D95
 
 | |
5LS6
 
 | | Structure of Human Polycomb Repressive Complex 2 (PRC2) with inhibitor | | 分子名称: | 1-[(1~{R})-1-[1-[2,2-bis(fluoranyl)propyl]piperidin-4-yl]ethyl]-~{N}-[(4-methoxy-6-methyl-2-oxidanylidene-3~{H}-pyridin-3-yl)methyl]-2-methyl-indole-3-carboxamide, Histone-lysine N-methyltransferase EZH2,Histone-lysine N-methyltransferase EZH2,Histone-lysine N-methyltransferase EZH2, Jarid2 K116me3, ... | | 著者 | Zhang, Y, Justin, N, Chen, S, Wilson, J, Gamblin, S. | | 登録日 | 2016-08-22 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.47 Å) | | 主引用文献 | Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase I Clinical Trials for B-Cell Lymphomas.
J. Med. Chem., 59, 2016
|
|
5VHE
 
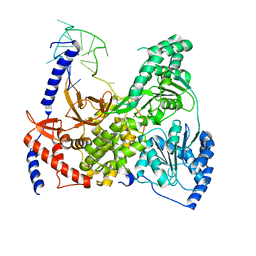 | | DHX36 in complex with the c-Myc G-quadruplex | | 分子名称: | DEAH (Asp-Glu-Ala-His) box polypeptide 36, DNA (5'-D(*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*TP*TP*TP*TP*TP*T)-3'), POTASSIUM ION | | 著者 | Chen, M, Ferre-D'Amare, A. | | 登録日 | 2017-04-13 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.793 Å) | | 主引用文献 | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5VHD
 
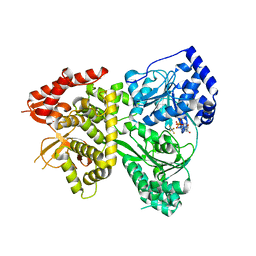 | |
5VHA
 
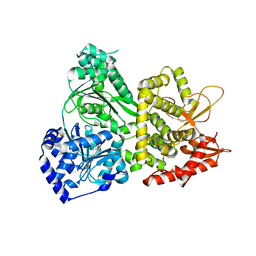 | |
5VHC
 
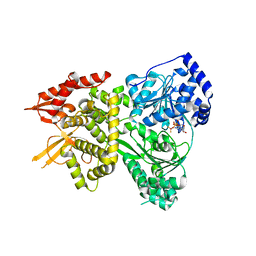 | |
8XV8
 
 | | Crystal structure of PHD domain of UHRF1 in complex with hStella peptide (residues 75-121) | | 分子名称: | Developmental pluripotency-associated protein 3, E3 ubiquitin-protein ligase UHRF1, ZINC ION | | 著者 | Du, X, Gan, Q, Xu, J, Liu, J. | | 登録日 | 2024-01-14 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Defining ortholog-specific UHRF1 inhibition by STELLA for cancer therapy.
Nat Commun, 16, 2025
|
|
8XV4
 
 | | Crystal structure of TTD-PHD domain of UHRF1 in complex with mStella peptide (residues 85-119) | | 分子名称: | Developmental pluripotency-associated protein 3, E3 ubiquitin-protein ligase UHRF1, ZINC ION | | 著者 | Du, X, Gan, Q, Xu, J, Liu, J. | | 登録日 | 2024-01-14 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-02-19 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Defining ortholog-specific UHRF1 inhibition by STELLA for cancer therapy.
Nat Commun, 16, 2025
|
|
8XV6
 
 | | Crystal structure of PHD domain of UHRF1 in complex with mStella peptide (residues 85-119) | | 分子名称: | Developmental pluripotency-associated protein 3, E3 ubiquitin-protein ligase UHRF1, GLYCEROL, ... | | 著者 | Du, X, Gan, Q, Xu, J, Liu, J. | | 登録日 | 2024-01-14 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Defining ortholog-specific UHRF1 inhibition by STELLA for cancer therapy.
Nat Commun, 16, 2025
|
|
8XV7
 
 | | Crystal structure of TTD-PHD domain of UHRF1 in complex with hStella peptide (residues 75-121) | | 分子名称: | ACETIC ACID, Developmental pluripotency-associated protein 3, E3 ubiquitin-protein ligase UHRF1, ... | | 著者 | Du, X, Gan, Q, Xu, J, Liu, J. | | 登録日 | 2024-01-14 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Defining ortholog-specific UHRF1 inhibition by STELLA for cancer therapy.
Nat Commun, 16, 2025
|
|
8E2M
 
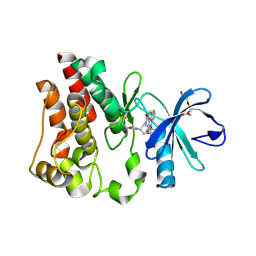 | | Bruton's tyrosine kinase (BTK) with compound 13 | | 分子名称: | (5P)-5-[4-methyl-6-(2-methylpropyl)pyridin-3-yl]-4-oxo-N-[(1R,2S)-2-propanamidocyclopentyl]-4,5-dihydro-3H-1-thia-3,5,8-triazaacenaphthylene-2-carboxamide, TRIETHYLENE GLYCOL, Tyrosine-protein kinase BTK | | 著者 | Alexander, R, Milligan, C.M. | | 登録日 | 2022-08-15 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.904 Å) | | 主引用文献 | Discovery of JNJ-64264681: A Potent and Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 65, 2022
|
|
3HS2
 
 | |
3HRY
 
 | |
7R7O
 
 | |
7R7C
 
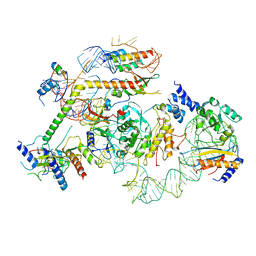 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - L1 stalk local model | | 分子名称: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-24 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R6K
 
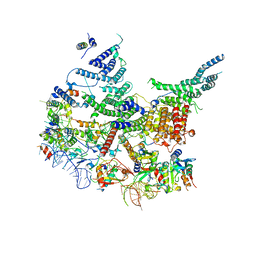 | | State E2 nucleolar 60S ribosomal intermediate - Model for Noc2/Noc3 region | | 分子名称: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-22 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R72
 
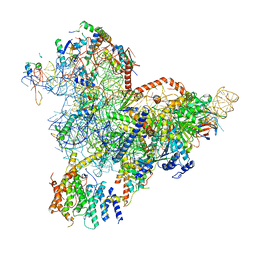 | | State E1 nucleolar 60S ribosome biogenesis intermediate - Spb4 local model | | 分子名称: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-24 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R6Q
 
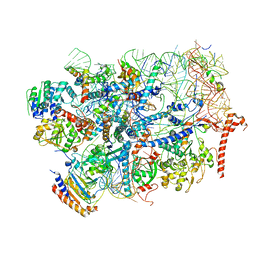 | | State E2 nucleolar 60S ribosome biogenesis intermediate - Foot region model | | 分子名称: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2021-06-23 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
