2IUF
 
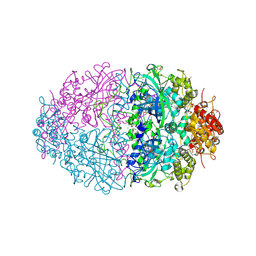 | | The structures of Penicillium vitale catalase: resting state, oxidised state (compound I) and complex with aminotriazole | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Murshudov, G, Borovik, A, Grebenko, A, Barynin, V, Vagin, A, Melik-Adamyan, W. | | 登録日 | 2006-06-02 | | 公開日 | 2006-07-10 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | The Structures and Electronic Configuration of Compound I Intermediates of Helicobacter Pylori and Penicillium Vitale Catalases Determined by X-Ray Crystallography and Qm/Mm Density Functional Theory Calculations.
J.Am.Chem.Soc., 129, 2007
|
|
2IQF
 
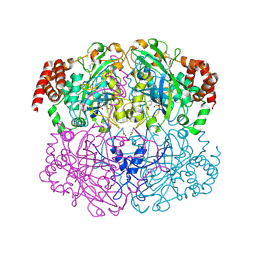 | | Crystal structure of Helicobacter pylori catalase compound I | | 分子名称: | ACETATE ION, Catalase, OXYGEN ATOM, ... | | 著者 | Loewen, P.C, Carpena, X, Fita, I. | | 登録日 | 2006-10-13 | | 公開日 | 2007-08-28 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | The structures and electronic configuration of compound I intermediates of Helicobacter pylori and Penicillium vitale catalases determined by X-ray crystallography and QM/MM density functional theory calculations.
J.Am.Chem.Soc., 129, 2007
|
|
8RVK
 
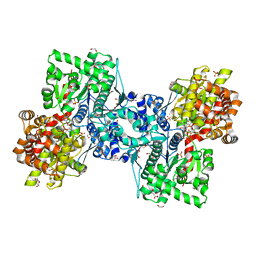 | | Maltodextrin phosphorylase (MalP) in complex with a alpha-1,2-cyclophellitol analogue | | 分子名称: | (3~{a}~{R},4~{R},5~{R},6~{R},7~{a}~{S})-6-(hydroxymethyl)-4,5-bis(oxidanyl)-3~{a},4,5,6,7,7~{a}-hexahydro-3~{H}-1,3-benzoxazol-2-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Bennett, M, Ofman, T.P, Overkleeft, H.S, Davies, G.J. | | 登録日 | 2024-02-01 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Conformational and Electronic Variations in 1,2- and 1,5a-Cyclophellitols and their Impact on Retaining alpha-Glucosidase Inhibition.
Chemistry, 30, 2024
|
|
8RZH
 
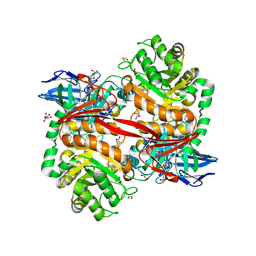 | | ZgGH129 from Zobellia galactanivorans in complex with the inhibitor AD-DGJ (3,6-anhydro-D-1-deoxygalactonojirimycin). | | 分子名称: | (1~{R},4~{S},5~{R},8~{S})-6-oxa-2-azabicyclo[3.2.1]octane-4,8-diol, 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, ... | | 著者 | Roret, T, Czjzek, M, Ficko-Blean, E. | | 登録日 | 2024-02-12 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RZJ
 
 | | ZgGH129 from Zobellia galactanivorans in complex with the inhibitor ADG-IF (3,6-anhydro-D-galacto-isofagomine). | | 分子名称: | (1~{R},5~{R},8~{S})-6-oxa-3-azabicyclo[3.2.1]octan-8-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Roret, T, Czjzek, M, Ficko-Blean, E. | | 登録日 | 2024-02-12 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RZI
 
 | | ZgGH129 from Zobellia galactanivorans soaked with 1,2-diF-ADG (3,6-Anhydro-2-deoxy-2-fluoro-a-D-galactopyranosyl fluoride) resulting in a trapped glycosyl-enzyme intermediate. | | 分子名称: | (1~{R},4~{S},5~{S},8~{S})-4-fluoranyl-2,6-dioxabicyclo[3.2.1]octan-8-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Roret, T, Czjzek, M, Ficko-Blean, E. | | 登録日 | 2024-02-12 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RZG
 
 | | ZgGH129 from Zobellia galactanivorans soaked with the product of the reaction ADG (3,6-anhydro-D-galactose). | | 分子名称: | (1~{R},4~{S},5~{R},8~{S})-2,6-dioxabicyclo[3.2.1]octane-4,8-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Roret, T, Czjzek, M, Ficko-Blean, E. | | 登録日 | 2024-02-12 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RZK
 
 | | The Michaelis complex of ZgGH129 D486N from Zobellia galactanivorans with neo-b/k-oligo-carrageenan tetrasaccharide (beta-kappa neo-oligo-carrageenan DP4). | | 分子名称: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CHLORIDE ION, ... | | 著者 | Roret, T, Czjzek, M, Ficko-Blean, E. | | 登録日 | 2024-02-12 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4JIE
 
 | | Structural analysis and insights into glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-mannosidase/beta-glucosidase, GLYCEROL, ... | | 著者 | Tankrathok, A, Luang, S, Robinson, R.C, Kimura, A, Hrmova, M, Ketudat Cairns, J.R. | | 登録日 | 2013-03-05 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural analysis and insights into the glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JHO
 
 | | Structural analysis and insights into glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-mannosidase/beta-glucosidase, GLYCEROL | | 著者 | Tankrathok, A, Luang, S, Robinson, R.C, Kimura, A, Hrmova, M, Ketudat Cairns, J.R. | | 登録日 | 2013-03-05 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structural analysis and insights into the glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6YJS
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with biantennary pentasaccharide M592 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, ... | | 著者 | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | 登録日 | 2020-04-04 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJQ
 
 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A,Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | 著者 | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | 登録日 | 2020-04-04 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJT
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | 著者 | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | 登録日 | 2020-04-04 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YQH
 
 | | GH146 beta-L-arabinofuranosidase bound to covalent inhibitor | | 分子名称: | (1~{S},2~{S},3~{S},4~{S})-4-(hydroxymethyl)cyclopentane-1,2,3-triol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetyl-CoA carboxylase, ... | | 著者 | McGregor, N.G.S, Davies, G.J. | | 登録日 | 2020-04-17 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6YJV
 
 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP-2-deoxy-2-fluoroglucose and biantennary pentasaccharide M592 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | 著者 | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | 登録日 | 2020-04-04 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJR
 
 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain. | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | 著者 | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | 登録日 | 2020-04-04 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJU
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP and biantennary pentasaccharide M592 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | 著者 | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | 登録日 | 2020-04-04 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
8R43
 
 | |
8QLI
 
 | |
8QMJ
 
 | |
8QIZ
 
 | | Crystal structure of Paradendryphiella salina PL7C alginate lyase mutant H124N soaked with with penta-mannuronic acid | | 分子名称: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | 著者 | Wilkens, C. | | 登録日 | 2023-09-12 | | 公開日 | 2025-02-05 | | 最終更新日 | 2025-07-02 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Unraveling the molecular mechanism of polysaccharide lyases for efficient alginate degradation.
Nat Commun, 16, 2025
|
|
9FHT
 
 | |
9FI4
 
 | |
9FI5
 
 | |
9FHU
 
 | |
