8X5Q
 
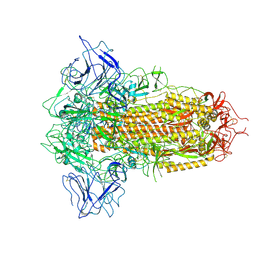 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-17 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8WHV
 
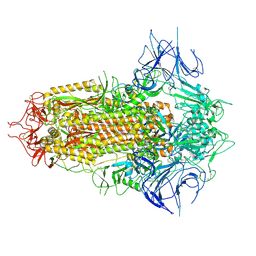 | | Spike Trimer of BA.2.86 with three RBDs down | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-09-23 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8X4Z
 
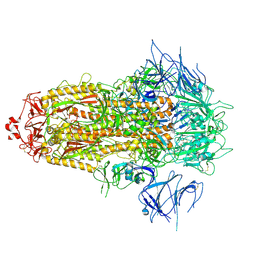 | | BA.2.86 Spike Trimer with ins483V mutation (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8XUS
 
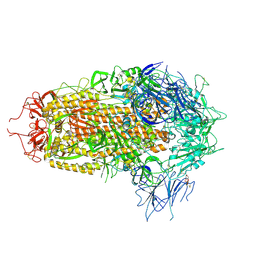 | | JN.1 Spike Trimer in complex with heparan sulfate | | 分子名称: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yue, C, Liu, P. | | 登録日 | 2024-01-14 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8WHS
 
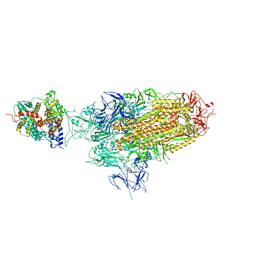 | | Spike Trimer of BA.2.86 in complex with one hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-09-23 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8X56
 
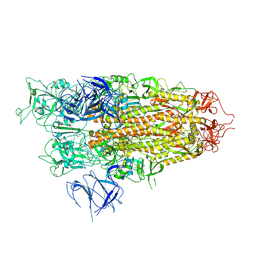 | | BA.2.86 Spike Trimer with T356K mutation (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8WHZ
 
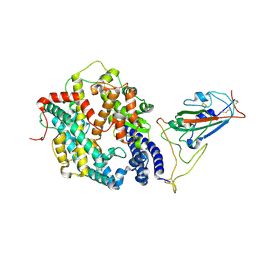 | | BA.2.86 RBD in complex with hACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-09-23 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8X55
 
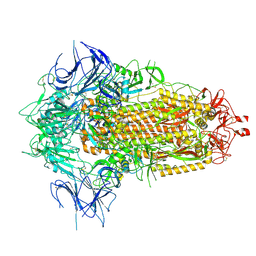 | | BA.2.86 Spike Trimer with T356K mutation (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
4TZC
 
 | | Crystal Structure of Murine Cereblon in Complex with Thalidomide | | 分子名称: | Protein cereblon, S-Thalidomide, SULFATE ION, ... | | 著者 | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | 登録日 | 2014-07-10 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
4TZU
 
 | | Crystal Structure of Murine Cereblon in Complex with Pomalidomide | | 分子名称: | Protein cereblon, S-Pomalidomide, SULFATE ION, ... | | 著者 | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | 登録日 | 2014-07-10 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
7VS3
 
 | | The crystal structure of rat calcium-dependent activator protein for secretion (CAPS) C2PH | | 分子名称: | Calcium-dependent secretion activator 1, SULFATE ION | | 著者 | Zhou, H, Wei, Z.Q, Zhang, L, Ren, Y.J, Ma, C. | | 登録日 | 2021-10-25 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.595 Å) | | 主引用文献 | The C 2 and PH domains of CAPS constitute an effective PI(4,5)P2-binding unit essential for Ca 2+ -regulated exocytosis.
Structure, 31, 2023
|
|
6WRO
 
 | |
6WRY
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE INPP1 IN COMPLEX GADOLINIUM AFTER ADDITION OF INOSITOL 1,3,4-TRISPHOSPHATE AT 2.5 ANGSTROM RESOLUTION | | 分子名称: | GADOLINIUM ATOM, Inositol polyphosphate 1-phosphatase, SULFATE ION | | 著者 | Dollins, D.R, Endo-Streeter, S, York, J.D. | | 登録日 | 2020-04-30 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
6WRR
 
 | |
6WE3
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with compound 3 | | 分子名称: | 1,2-ETHANEDIOL, 2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}-8-methylquinazolin-4(3H)-one, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Swinger, K.S, Schenkel, L.B, Kuntz, K.W. | | 登録日 | 2020-04-01 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
6WE2
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN012759 | | 分子名称: | 1,2-ETHANEDIOL, 7-(cyclopropylmethoxy)-5-fluoro-2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}quinazolin-4(3H)-one, Isoform 1 of Protein mono-ADP-ribosyltransferase PARP14 | | 著者 | Swinger, K.K, Schenkel, L.B, Kuntz, K.W. | | 登録日 | 2020-04-01 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
6WE4
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with compound 2 | | 分子名称: | 1,2-ETHANEDIOL, 2-methyl-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one, 8-methyl-2-{[(pyridin-4-yl)sulfanyl]methyl}quinazolin-4(3H)-one, ... | | 著者 | Swinger, K.S, Schenkel, L.B, Kuntz, K.W. | | 登録日 | 2020-04-01 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
8JMT
 
 | | Structure of the adhesion GPCR ADGRL3 in the apo state | | 分子名称: | Adhesion G protein-coupled receptor L3,Soluble cytochrome b562 | | 著者 | Tao, Y, Guo, Q, He, B, Zhong, Y. | | 登録日 | 2023-06-05 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8J8Q
 
 | | Structure of the four-component Paf1 complex from Saccharomyces eubayanus | | 分子名称: | CDC73-like protein, CTR9-like protein, PAF1-like protein, ... | | 著者 | Wang, Z, Qin, Y, Zhou, Y, Cao, Y. | | 登録日 | 2023-05-02 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Structural Basis of the Transcriptional Elongation Factor Paf1 Core Complex from Saccharomyces eubayanus .
Int J Mol Sci, 24, 2023
|
|
8JJ8
 
 | | Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with a partial agonist | | 分子名称: | Beta-2 adrenergic receptor,Soluble cytochrome b562, ~{N}-[5-[(1~{R})-2-[[(2~{R})-1-(4-methoxyphenyl)propan-2-yl]amino]-1-oxidanyl-ethyl]-2-oxidanyl-phenyl]methanamide | | 著者 | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | 登録日 | 2023-05-29 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8JJL
 
 | | cryo-EM structure of the beta2-AR-mBRIL/1b3 Fab/Glue complex with a full agonist | | 分子名称: | Beta-2 adrenergic receptor,Soluble cytochrome b562, Olodaterol | | 著者 | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | 登録日 | 2023-05-30 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8J7E
 
 | |
8JJO
 
 | | Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with an antagonist | | 分子名称: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, Beta-2 adrenergic receptor,Beta-2 adrenergic receptor,Soluble cytochrome b562 | | 著者 | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | 登録日 | 2023-05-31 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8J8P
 
 | | Structure of the four-component Paf1 complex from Saccharomyces eubayanus | | 分子名称: | CDC73-like protein, CTR9-like protein, PAF1-like protein, ... | | 著者 | Wang, Z, Qin, Y, Zhou, Y, Cao, Y. | | 登録日 | 2023-05-02 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Basis of the Transcriptional Elongation Factor Paf1 Core Complex from Saccharomyces eubayanus .
Int J Mol Sci, 24, 2023
|
|
8JBA
 
 | |
