7BXR
 
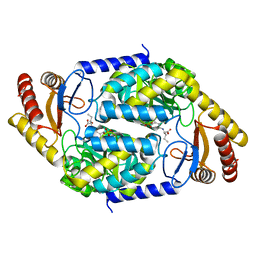 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator 3-Hydroxynorvaline binding form | | 分子名称: | (2S,3R)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-pentanoic acid, 2-amino-3-ketobutyrate coenzyme A ligase | | 著者 | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | 登録日 | 2020-04-20 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7CCU
 
 | |
7CCV
 
 | |
7CCW
 
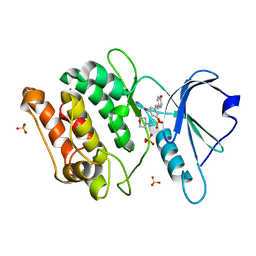 | | Crystal structure of death-associated protein kinase 1 in complex with resveratrol and MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Death-associated protein kinase 1, RESVERATROL, ... | | 著者 | Yokoyama, T, Suzuki, R, Mizuguchi, M. | | 登録日 | 2020-06-18 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of death-associated protein kinase 1 in complex with the dietary compound resveratrol.
Iucrj, 8, 2020
|
|
7F4Z
 
 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 1B, ... | | 著者 | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | 登録日 | 2021-06-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
7F4X
 
 | | Joint neutron and X-ray crystal structure of the nucleotide-binding domain of Hsp72 in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1B, MAGNESIUM ION, ... | | 著者 | Yokoyama, T, Ostermann, A, Schrader, T.E. | | 登録日 | 2021-06-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | 主引用文献 | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
7F50
 
 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with AMPPnP | | 分子名称: | CHLORIDE ION, Heat shock 70 kDa protein 1B, MAGNESIUM ION, ... | | 著者 | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | 登録日 | 2021-06-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.703 Å) | | 主引用文献 | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
7CGV
 
 | | Full consensus L-threonine 3-dehydrogenase, FcTDH-IIYM (NAD+ bound form) | | 分子名称: | Artificial L-threonine 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Motoyama, T, Hiramatsu, N, Asano, Y, Nakano, S, Ito, S. | | 登録日 | 2020-07-02 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Protein Sequence Selection Method That Enables Full Consensus Design of Artificial l-Threonine 3-Dehydrogenases with Unique Enzymatic Properties.
Biochemistry, 59, 2020
|
|
8Z56
 
 | |
8Z55
 
 | |
8Z54
 
 | |
8Z58
 
 | |
8Z57
 
 | |
7X7K
 
 | | Ancestral L-Lys oxidase (AncLLysO-2) L-Arg binding form | | 分子名称: | ARGININE, FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | 登録日 | 2022-03-09 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
7X7J
 
 | | Ancestral L-Lys oxidase (AncLLysO-2) L-Lys binding form | | 分子名称: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE | | 著者 | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | 登録日 | 2022-03-09 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
7X7I
 
 | | Ancestral L-Lys oxidase (AncLLysO-2) ligand free form | | 分子名称: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | 登録日 | 2022-03-09 | | 公開日 | 2023-01-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
3BNV
 
 | |
3D6L
 
 | |
1V4U
 
 | | Crystal structure of bluefin tuna carbonmonoxy-hemoglobin | | 分子名称: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha chain, ... | | 著者 | Yokoyama, T, Chong, K.T, Miyazaki, Y, Nakatsukasa, T, Unzai, S, Miyazaki, G, Morimoto, H, Jeremy, R.H.T, Park, S.Y. | | 登録日 | 2003-11-19 | | 公開日 | 2004-07-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Novel Mechanisms of pH Sensitivity in Tuna Hemoglobin: A STRUCTURAL EXPLANATION OF THE ROOT EFFECT
J.Biol.Chem., 279, 2004
|
|
1V4X
 
 | | Crystal structure of bluefin tuna hemoglobin deoxy form at pH5.0 | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha chain, hemoglobin beta chain | | 著者 | Yokoyama, T, Chong, K.T, Miyazaki, Y, Nakatsukasa, T, Unzai, S, Miyazaki, G, Morimoto, H, Jeremy, R.H.T, Park, S.Y. | | 登録日 | 2003-11-19 | | 公開日 | 2004-07-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Novel Mechanisms of pH Sensitivity in Tuna Hemoglobin: A STRUCTURAL EXPLANATION OF THE ROOT EFFECT
J.Biol.Chem., 279, 2004
|
|
1V4W
 
 | | Crystal structure of bluefin tuna hemoglobin deoxy form at pH7.5 | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha chain, hemoglobin beta chain | | 著者 | Yokoyama, T, Chong, K.T, Miyazaki, Y, Nakatsukasa, T, Unzai, S, Miyazaki, G, Morimoto, H, Jeremy, R.H.T, Park, S.Y. | | 登録日 | 2003-11-19 | | 公開日 | 2004-07-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Novel Mechanisms of pH Sensitivity in Tuna Hemoglobin: A STRUCTURAL EXPLANATION OF THE ROOT EFFECT
J.Biol.Chem., 279, 2004
|
|
2D5Z
 
 | | Crystal structure of T-state human hemoglobin complexed with three L35 molecules | | 分子名称: | 2-[4-({[(3,5-DICHLOROPHENYL)AMINO]CARBONYL}AMINO)PHENOXY]-2-METHYLPROPANOIC ACID, Hemoglobin alpha subunit, Hemoglobin beta subunit, ... | | 著者 | Yokoyama, T, Neya, S, Tsuneshige, A, Yonetani, T, Park, S.Y, Tame, J.R. | | 登録日 | 2005-11-08 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | R-state haemoglobin with low oxygen affinity: crystal structures of deoxy human and carbonmonoxy horse haemoglobin bound to the effector molecule L35
J.Mol.Biol., 356, 2006
|
|
2D60
 
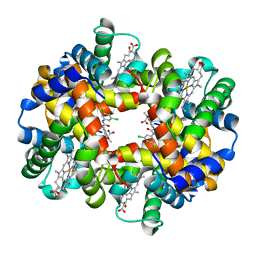 | | Crystal structure of deoxy human hemoglobin complexed with two L35 molecules | | 分子名称: | 2-[4-({[(3,5-DICHLOROPHENYL)AMINO]CARBONYL}AMINO)PHENOXY]-2-METHYLPROPANOIC ACID, Hemoglobin alpha subunit, Hemoglobin beta subunit, ... | | 著者 | Yokoyama, T, Neya, S, Tsuneshige, A, Yonetani, T, Park, S.Y, Tame, J.R. | | 登録日 | 2005-11-08 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | R-state haemoglobin with low oxygen affinity: crystal structures of deoxy human and carbonmonoxy horse haemoglobin bound to the effector molecule L35
J.Mol.Biol., 356, 2006
|
|
2D5X
 
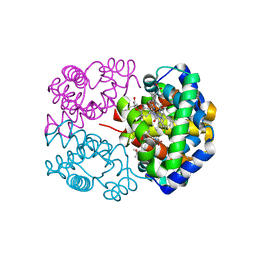 | | Crystal structure of carbonmonoxy horse hemoglobin complexed with L35 | | 分子名称: | 2-[4-({[(3,5-DICHLOROPHENYL)AMINO]CARBONYL}AMINO)PHENOXY]-2-METHYLPROPANOIC ACID, CARBON MONOXIDE, Hemoglobin alpha subunit, ... | | 著者 | Yokoyama, T, Neya, S, Tsuneshige, A, Yonetani, T, Park, S.Y, Tame, J.R. | | 登録日 | 2005-11-08 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | R-state haemoglobin with low oxygen affinity: crystal structures of deoxy human and carbonmonoxy horse haemoglobin bound to the effector molecule L35
J.Mol.Biol., 356, 2006
|
|
4YT1
 
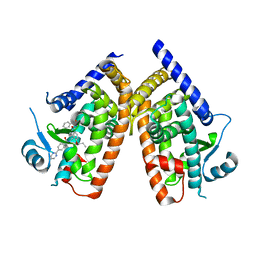 | | Human PPAR Gamma Ligand Binding Domain in complex with a Gammma Selective Synthetic Partial Agonist MEKT76 | | 分子名称: | N-(benzylsulfonyl)-4-propoxy-3-({[4-(pyrimidin-2-yl)benzoyl]amino}methyl)benzamide, Peroxisome proliferator-activated receptor gamma | | 著者 | Oyama, T, Ohashi, M, Miyachi, H, Kusunoki, M. | | 登録日 | 2015-03-17 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Peroxisome proliferator-activated receptor gamma (PPAR gamma ) has multiple binding points that accommodate ligands in various conformations: Structurally similar PPAR gamma partial agonists bind to PPAR gamma LBD in different conformations
Bioorg.Med.Chem.Lett., 25, 2015
|
|
