1GJA
 
 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | 分子名称: | CITRIC ACID, N-(4-CARBAMIMIDOYL-PHENYL)-2-HYDROXY-BENZAMIDE, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | 著者 | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | 登録日 | 2001-04-27 | | 公開日 | 2002-04-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GJ5
 
 | | SELECTIVITY AT S1, H2O DISPLACEMENT, UPA, TPA, SER190/ALA190 PROTEASE, STRUCTURE-BASED DRUG DESIGN | | 分子名称: | 2-(2-HYDROXY-BIPHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, SODIUM ION, ... | | 著者 | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | 登録日 | 2001-04-27 | | 公開日 | 2002-04-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GJB
 
 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | 分子名称: | 2-(2-HYDROXY-BIPHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | 著者 | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | 登録日 | 2001-04-27 | | 公開日 | 2002-04-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GHX
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | 分子名称: | 2-(2-HYDROXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, CALCIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | 登録日 | 2001-01-22 | | 公開日 | 2002-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI7
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | 分子名称: | 2-(2-OXO-1,2-DIHYDRO-PYRIDIN-3-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | 著者 | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | 登録日 | 2001-01-22 | | 公開日 | 2002-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GJ7
 
 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | 分子名称: | 6-CHLORO-2-(2-HYDROXY-BIPHENYL-3-YL)-1H-INDOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | 著者 | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | 登録日 | 2001-04-27 | | 公開日 | 2002-04-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GHY
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | 分子名称: | 2-(3-HYDROXY-PYRIDIN-2-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, CALCIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | 登録日 | 2001-01-22 | | 公開日 | 2002-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GJ9
 
 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | 分子名称: | 6-FLUORO-2-[2-HYDROXY-3-(2-METHYL-CYCLOHEXYLOXY)-PHENYL]-1H-INDOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | 著者 | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | 登録日 | 2001-04-30 | | 公開日 | 2002-04-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GHV
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | 分子名称: | 2-(2-OXO-1,2-DIHYDRO-PYRIDIN-3-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, SODIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | 登録日 | 2001-01-22 | | 公開日 | 2002-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI9
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | 分子名称: | 2-(2-HYDROXY-5-METHOXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | 著者 | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | 登録日 | 2001-01-22 | | 公開日 | 2002-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI2
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | 分子名称: | 2-(2-HYDROXY-PHENYL)-3H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | 登録日 | 2001-01-22 | | 公開日 | 2002-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1C1T
 
 | |
1C2L
 
 | |
1C1Q
 
 | |
1C1P
 
 | |
1C2M
 
 | |
1C2G
 
 | |
1C1N
 
 | |
1O2G
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | 分子名称: | 3-{5-[AMINO(IMINIO)METHYL]-1H-INDOL-2-YL}-1,1'-BIPHENYL-2-OLATE, ACETYL HIRUDIN, SODIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | 登録日 | 2003-03-06 | | 公開日 | 2003-05-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
1O2J
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | 分子名称: | 2-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-6-ISOBUTOXYBENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | 登録日 | 2003-03-06 | | 公開日 | 2003-05-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
1O2R
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | 分子名称: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | 登録日 | 2003-03-06 | | 公開日 | 2003-09-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
1O2Z
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | 分子名称: | 2-(5-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-2'-METHOXY-6-OXIDO-1,1'-BIPHENYL-3-YL)SUCCINATE, BETA-TRYPSIN, CALCIUM ION | | 著者 | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | 登録日 | 2003-03-06 | | 公開日 | 2003-09-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
1O35
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | 分子名称: | 2-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-4-FLUOROBENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | 登録日 | 2003-03-06 | | 公開日 | 2003-09-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
1O3C
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | 分子名称: | 3-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-1,1'-BIPHENYL-2-OLATE, BETA-TRYPSIN, CALCIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | 登録日 | 2003-03-06 | | 公開日 | 2003-09-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
1O3K
 
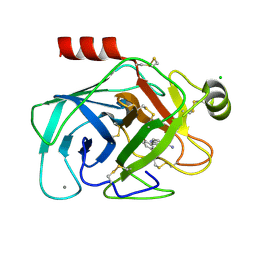 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | 分子名称: | 2-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-6-BROMO-4-METHYLBENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | 登録日 | 2003-03-06 | | 公開日 | 2003-09-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
