2IPC
 
 | | Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer | | 分子名称: | Preprotein translocase SecA subunit | | 著者 | Vassylyev, D.G, Mori, H, Vassylyeva, M.N, Tsukazaki, T, Kimura, Y, Tahirov, T.H, Ito, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-10-12 | | 公開日 | 2006-11-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of the Translocation ATPase SecA from Thermus thermophilus Reveals a Parallel, Head-to-Head Dimer.
J.Mol.Biol., 364, 2006
|
|
1MBQ
 
 | | Anionic Trypsin from Pacific Chum Salmon | | 分子名称: | BENZAMIDINE, CALCIUM ION, Trypsin | | 著者 | Toyota, E, Ng, K.K.S, Kuninaga, S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | 登録日 | 2002-08-03 | | 公開日 | 2002-12-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Nucleotide Sequence of an Anionic Trypsin from Chum Salmon (Oncorhynchus keta) in Comparison with Atlantic Salmon (Salmo salar) and Bovine Trypsin
J.Mol.Biol., 324, 2002
|
|
5YT0
 
 | | Crystal structure of the complex of archaeal ribosomal stalk protein aP1 and archaeal translation initiation factor aIF5B | | 分子名称: | Archaeal ribosomal stalk protein aP1, GUANOSINE-5'-DIPHOSPHATE, Probable translation initiation factor IF-2 | | 著者 | Murakami, R, Singh, C.R, Morris, J, Tang, L, Harmon, I, Miyoshi, T, Ito, K, Asano, K, Uchiumi, T. | | 登録日 | 2017-11-16 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | The Interaction between the Ribosomal Stalk Proteins and Translation Initiation Factor 5B Promotes Translation Initiation
Mol. Cell. Biol., 38, 2018
|
|
6KB2
 
 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by soaking | | 分子名称: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KAZ
 
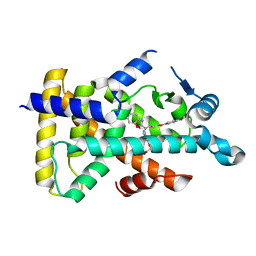 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by soaking | | 分子名称: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KBA
 
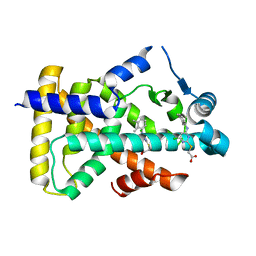 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by co-crystallization | | 分子名称: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KAX
 
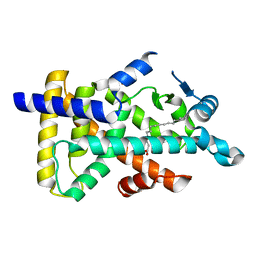 | | X-ray structure of human PPARalpha ligand binding domain-intrinsic fatty acid (E. coli origin) co-crystals obtained by cross-seeding | | 分子名称: | GLYCEROL, PALMITIC ACID, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB9
 
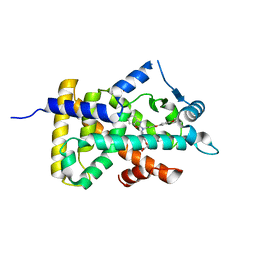 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by cross-seeding | | 分子名称: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB5
 
 | | X-ray structure of human PPARalpha ligand binding domain-5,8,11,14-eicosatetraynoic Acid (ETYA) co-crystals obtained by delipidation and cross-seeding | | 分子名称: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, icosa-5,8,11,14-tetraynoic acid | | 著者 | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB4
 
 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by delipidation and cross-seeding | | 分子名称: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB6
 
 | | X-ray structure of human PPARalpha ligand binding domain-tetradecylthioacetic acid (TTA) co-crystals obtained by delipidation and cross-seeding | | 分子名称: | 2-tetradecylsulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.431 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7COH
 
 | | Dimeric Form of Bovine Heart Cytochrome c Oxidase in the Fully Oxidized State | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | 著者 | Shinzawa-Itoh, K, Muramoto, K. | | 登録日 | 2020-08-04 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The 1.3-A Resolution structure of bovine cytochrome c oxidase suggests a dimerization mechanism
Biochim.Biophys.Acta, 2021
|
|
5YV5
 
 | | Crystal structure of the complex of archaeal ribosomal stalk protein aP1 and archaeal ribosome recycling factor aABCE1. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATPase RIL, Archaeal ribosomal stalk protein aP1, ... | | 著者 | Imai, H, Abe, T, Miyoshi, T, Nishikawa, S, Ito, K, Uchiumi, T. | | 登録日 | 2017-11-24 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The ribosomal stalk protein is crucial for the action of the conserved ATPase ABCE1
Nucleic Acids Res., 46, 2018
|
|
6JI2
 
 | | Crystal structure of archaeal ribosomal protein aP1, aPelota, and GTP-bound aEF1A complex | | 分子名称: | Archaeal ribosomal stalk protein aP1, Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Maruyama, K, Imai, H, Kawamura, M, Ishino, S, Ishino, Y, Ito, K, Uchiumi, T. | | 登録日 | 2019-02-20 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Switch of the interactions between the ribosomal stalk and EF1A in the GTP- and GDP-bound conformations.
Sci Rep, 9, 2019
|
|
1UGP
 
 | | Crystal structure of Co-type nitrile hydratase complexed with n-butyric acid | | 分子名称: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, ... | | 著者 | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | 登録日 | 2003-06-17 | | 公開日 | 2004-06-17 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGS
 
 | | Crystal structure of aY114T mutant of Co-type nitrile hydratase | | 分子名称: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | 著者 | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | 登録日 | 2003-06-17 | | 公開日 | 2004-06-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGR
 
 | | Crystal structure of aT109S mutant of Co-type nitrile hydratase | | 分子名称: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | 著者 | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | 登録日 | 2003-06-17 | | 公開日 | 2004-06-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGQ
 
 | | Crystal structure of apoenzyme of Co-type nitrile hydratase | | 分子名称: | Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | 著者 | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | 登録日 | 2003-06-17 | | 公開日 | 2004-06-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1V7Z
 
 | | creatininase-product complex | | 分子名称: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | 著者 | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | 登録日 | 2003-12-26 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1V3Y
 
 | | The crystal structure of peptide deformylase from Thermus thermophilus HB8 | | 分子名称: | Peptide deformylase | | 著者 | Kamo, M, Kudo, N, Lee, W.C, Ito, K, Motoshim, H, Tanokura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-11-07 | | 公開日 | 2004-12-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | The crystal structure of peptide deformylase from Thermus thermophilus HB8
to be published
|
|
5FG3
 
 | | Crystal structure of GDP-bound aIF5B from Aeropyrum pernix | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Probable translation initiation factor IF-2 | | 著者 | Murakami, R, Miyoshi, T, Uchiumi, T, Ito, K. | | 登録日 | 2015-12-20 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of translation initiation factor 5B from the crenarchaeon Aeropyrum pernix.
Proteins, 84, 2016
|
|
6J3L
 
 | | Solution structure of the N-terminal extended protuberant domain of eukaryotic ribosomal stalk protein P0 | | 分子名称: | 60S acidic ribosomal protein P0 | | 著者 | Choi, K.H.A, Lee, K.M, Yang, L, Wing-Heng Yu, C, Banfield, D.K, Ito, K, Uchiumi, T, Wong, K.B. | | 登録日 | 2019-01-04 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Mutagenesis Studies Evince the Role of the Extended Protuberant Domain of Ribosomal Protein uL10 in Protein Translation.
Biochemistry, 58, 2019
|
|
5ZX8
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Thermus thermophilus | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRATE ANION, Peptidyl-tRNA hydrolase | | 著者 | Matsumoto, A, Uehara, U, Shimizu, Y, Ueda, T, Uchiumi, T, Ito, K. | | 登録日 | 2018-05-18 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | High-resolution crystal structure of peptidyl-tRNA hydrolase from Thermus thermophilus.
Proteins, 87, 2019
|
|
1IRE
 
 | | Crystal Structure of Co-type nitrile hydratase from Pseudonocardia thermophila | | 分子名称: | COBALT (II) ION, Nitrile Hydratase | | 著者 | Miyanaga, A, Fushinobu, S, Ito, K, Wakagi, T. | | 登録日 | 2001-10-01 | | 公開日 | 2002-10-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of cobalt-containing nitrile hydratase.
Biochem.Biophys.Res.Commun., 288, 2001
|
|
2CZK
 
 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) (trigonal form) | | 分子名称: | Inositol monophosphatase 2 | | 著者 | Arai, R, Ito, K, Hanawa-Suetsugu, K, Ohnishi, T, Ohba, H, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-07-13 | | 公開日 | 2006-07-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
