6DCZ
 
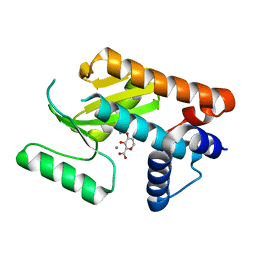 | |
6DFH
 
 | | BG505 MD64 N332-GT2 SOSIP trimer in complex with germline-reverted BG18 fragment antigen binding | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | 著者 | Ozorowski, G, Steichen, J.M, Schief, W.R, Ward, A.B. | | 登録日 | 2018-05-14 | | 公開日 | 2019-11-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
6DCY
 
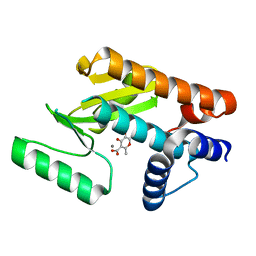 | |
6DMX
 
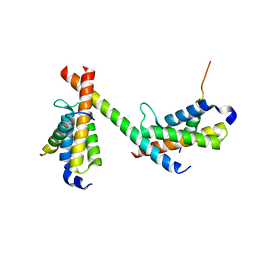 | | HBZ56 in complex with KIX and c-Myb | | 分子名称: | BZIP factor, CREB-binding protein, Transcriptional activator Myb | | 著者 | Yang, K, Wright, P.E, Stanfield, R.L. | | 登録日 | 2018-06-05 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DNQ
 
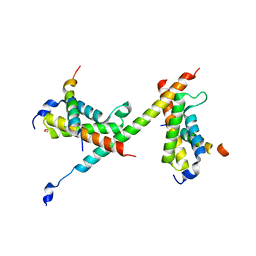 | | HBZ77 in complex with KIX and c-Myb | | 分子名称: | 1,2-ETHANEDIOL, BZIP factor, CREB-binding protein, ... | | 著者 | Yang, K, Wright, P.E, Stanfield, R.L. | | 登録日 | 2018-06-07 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DCQ
 
 | | Ectodomain of full length, wild type HIV-1 glycoprotein clone PC64M18C043 in complex with PGT151 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Rantalainen, K. | | 登録日 | 2018-05-08 | | 公開日 | 2018-06-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Co-evolution of HIV Envelope and Apex-Targeting Neutralizing Antibody Lineage Provides Benchmarks for Vaccine Design.
Cell Rep, 23, 2018
|
|
6DFG
 
 | | BG505 MD39 SOSIP trimer in complex with mature BG18 fragment antigen binding | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | 著者 | Ozorowski, G, Steichen, J.M, Schief, W.R, Ward, A.B. | | 登録日 | 2018-05-14 | | 公開日 | 2019-11-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.42 Å) | | 主引用文献 | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
6E0Q
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 4,6-dihydroxy-2-methyl-5-oxocyclohepta-1,3,6-triene-1-carboxylic acid | | 分子名称: | 1,2-ETHANEDIOL, 4,6-dihydroxy-2-methyl-5-oxocyclohepta-1,3,6-triene-1-carboxylic acid, MANGANESE (II) ION, ... | | 著者 | Dick, B.L, Morrison, C.N, Cohen, S.M. | | 登録日 | 2018-07-06 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure-Activity Relationships in Metal-Binding Pharmacophores for Influenza Endonuclease.
J. Med. Chem., 61, 2018
|
|
6DZQ
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 3-hydroxy-6-methyl-4-oxo-4H-pyran-2-carboxylic acid | | 分子名称: | 1,2-ETHANEDIOL, 3-hydroxy-6-methyl-4-oxo-4H-pyran-2-carboxylic acid, MANGANESE (II) ION, ... | | 著者 | Dick, B.L, Morrison, C.N, Cohen, S.M. | | 登録日 | 2018-07-05 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-Activity Relationships in Metal-Binding Pharmacophores for Influenza Endonuclease.
J. Med. Chem., 61, 2018
|
|
2KYS
 
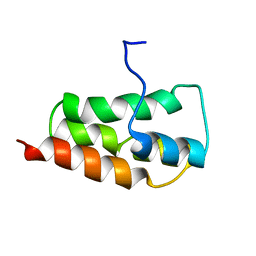 | | NMR Structure of the SARS Coronavirus Nonstructural Protein Nsp7 in Solution at pH 6.5 | | 分子名称: | Non-structural protein 7 | | 著者 | Johnson, M.A, Jaudzems, K, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2010-06-07 | | 公開日 | 2010-06-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of the SARS-CoV Nonstructural Protein 7 in Solution at pH 6.5.
J.Mol.Biol., 402, 2010
|
|
2JZD
 
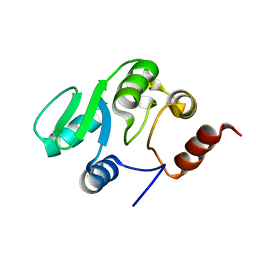 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3 | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2008-01-04 | | 公開日 | 2008-02-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZE
 
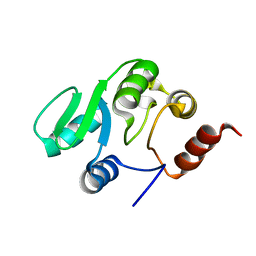 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3, single conformer closest to the mean coordinates of an ensemble of twenty energy minimized conformers | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2008-01-04 | | 公開日 | 2008-02-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZF
 
 | | NMR Conformer closest to the mean coordinates of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2008-01-04 | | 公開日 | 2008-02-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2KAF
 
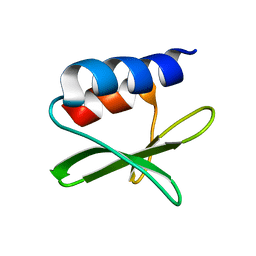 | | Solution structure of the SARS-unique domain-C from the nonstructural protein 3 (nsp3) of the severe acute respiratory syndrome coronavirus | | 分子名称: | Non-structural protein 3 | | 著者 | Johnson, M.A, Mohanty, B, Pedrini, B, Serrano, P, Chatterjee, A, Herrmann, T, Joseph, J, Saikatendu, K, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2008-11-05 | | 公開日 | 2008-11-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding.
J.Mol.Biol., 400, 2010
|
|
4EPS
 
 | |
4GEZ
 
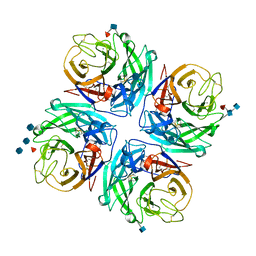 | | Structure of a neuraminidase-like protein from A/bat/Guatemala/164/2009 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Yang, H, Carney, P.J, Donis, R.O, Stevens, J. | | 登録日 | 2012-08-02 | | 公開日 | 2012-09-26 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DGU
 
 | |
4FDY
 
 | |
4H40
 
 | |
4H4J
 
 | |
4HMG
 
 | | REFINEMENT OF THE INFLUENZA VIRUS HEMAGGLUTININ BY SIMULATED ANNEALING | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | 著者 | Weis, W.I, Bruenger, A.T, Skehel, J.J, Wiley, D.C. | | 登録日 | 1989-09-11 | | 公開日 | 1991-01-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Refinement of the influenza virus hemagglutinin by simulated annealing.
J.Mol.Biol., 212, 1990
|
|
4GPV
 
 | |
1EO8
 
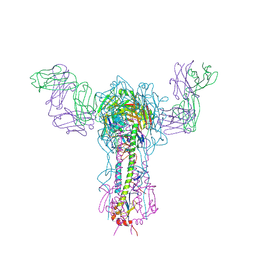 | | INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH A NEUTRALIZING ANTIBODY | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY (HEAVY CHAIN), ... | | 著者 | Fleury, D, Gigant, B, Daniels, R.S, Skehel, J.J, Knossow, M, Bizebard, T. | | 登録日 | 2000-03-22 | | 公開日 | 2000-04-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural evidence for recognition of a single epitope by two distinct antibodies.
Proteins, 40, 2000
|
|
4M61
 
 | |
3JXO
 
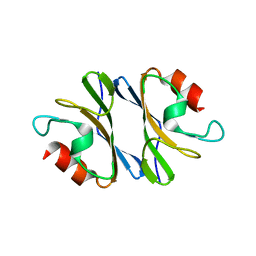 | | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima | | 分子名称: | TrkA-N domain protein | | 著者 | Deller, M.C, Johnson, H.A, Miller, M, Spraggon, G, Wilson, I.A, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2009-09-20 | | 公開日 | 2009-10-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima
To be Published
|
|
