4FB5
 
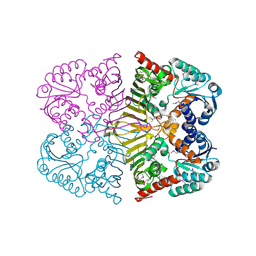 | |
1S8L
 
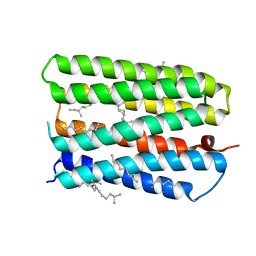 | | Anion-free form of the D85S mutant of bacteriorhodopsin from crystals grown in the presence of halide | | 分子名称: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin precursor, RETINAL | | 著者 | Facciotti, M.T, Cheung, V.S, Lunde, C.S, Rouhani, S, Baliga, N.S, Glaeser, R.M. | | 登録日 | 2004-02-02 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Specificity of anion binding in the substrate pocket of bacteriorhodopsin.
Biochemistry, 43, 2004
|
|
3KTN
 
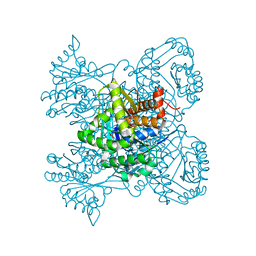 | |
3KZH
 
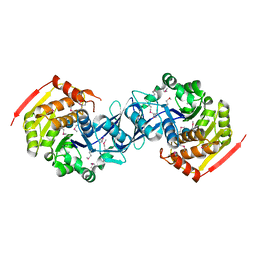 | |
6ISC
 
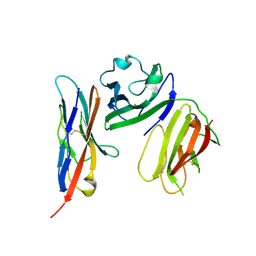 | | complex structure of mCD226-ecto and hCD155-D1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, Poliovirus receptor | | 著者 | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | 登録日 | 2018-11-16 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3JYI
 
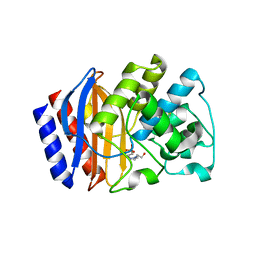 | | Structural and biochemical evidence that a TEM-1 {beta}-lactamase Asn170Gly active site mutant acts via substrate-assisted catalysis | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase TEM, PHOSPHATE ION | | 著者 | Brown, N.G, Palzkill, T.G, Prasad, B.V.V, Shanker, S. | | 登録日 | 2009-09-21 | | 公開日 | 2009-10-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.703 Å) | | 主引用文献 | Structural and biochemical evidence that a TEM-1 beta-lactamase N170G active site mutant acts via substrate-assisted catalysis
J.Biol.Chem., 284, 2009
|
|
1S8J
 
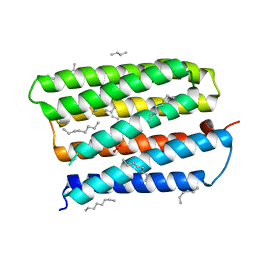 | | Nitrate-bound D85S mutant of bacteriorhodopsin | | 分子名称: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin precursor, NITRATE ION, ... | | 著者 | Facciotti, M.T, Cheung, V.S, Lunde, C.S, Rouhani, S, Baliga, N.S, Glaeser, R.M. | | 登録日 | 2004-02-02 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Specificity of anion binding in the substrate pocket of bacteriorhodopsin.
Biochemistry, 43, 2004
|
|
3BQX
 
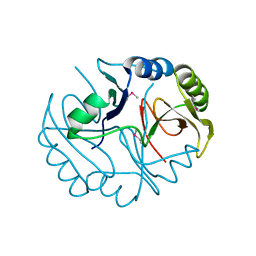 | |
3C8A
 
 | |
3KD9
 
 | |
3BBL
 
 | |
3HUU
 
 | |
3I3Y
 
 | |
3HV1
 
 | |
3BQT
 
 | |
3HYO
 
 | | Crystal structure of pyridoxal kinase from Lactobacillus plantarum in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Pyridoxal kinase | | 著者 | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-06-22 | | 公開日 | 2009-06-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of pyridoxal kinase from Lactobacillus plantarum in complex with ADP
To be Published
|
|
3HUT
 
 | |
3BZW
 
 | | Crystal structure of a putative lipase from Bacteroides thetaiotaomicron | | 分子名称: | ACETATE ION, Putative lipase, SULFATE ION | | 著者 | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-01-18 | | 公開日 | 2008-02-05 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Crystal structure of a putative lipase from Bacteroides thetaiotaomicron.
To be Published
|
|
1RTT
 
 | | Crystal structure determination of a putative NADH-dependent reductase using sulfur anomalous signal | | 分子名称: | SULFATE ION, conserved hypothetical protein | | 著者 | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-12-10 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Structure determination of an FMN reductase from Pseudomonas aeruginosa PA01 using sulfur anomalous signal.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
3I3V
 
 | |
1RVI
 
 | | SOLUTION STRUCTURE OF THE DNA DODECAMER CGTTTTAAAACG | | 分子名称: | 5'-D(*CP*GP*TP*TP*TP*TP*AP*AP*AP*AP*CP*G)-3' | | 著者 | Stefl, R, Wu, H, Ravindranathan, S, Sklenar, V, Feigon, J. | | 登録日 | 2003-12-13 | | 公開日 | 2004-02-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | DNA A-tract bending in three dimensions: Solving the dA4T4 vs. dT4A4 conundrum.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RW0
 
 | |
4RIS
 
 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | 分子名称: | CH58-UA Fab heavy chain, CH58-UA Fab light chain, Envelope glycoprotein | | 著者 | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | 登録日 | 2014-10-07 | | 公開日 | 2015-08-12 | | 最終更新日 | 2015-09-02 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
4RIR
 
 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | 分子名称: | CH58-UA Fab heavy chain, CH58-UA Fab light chain | | 著者 | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | 登録日 | 2014-10-07 | | 公開日 | 2015-08-12 | | 最終更新日 | 2015-09-02 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
3BGA
 
 | |
