1L01
 
 | |
1L07
 
 | |
1L08
 
 | |
1L04
 
 | |
1L05
 
 | |
1L02
 
 | |
1L09
 
 | |
3GOA
 
 | | Crystal structure of the Salmonella typhimurium FadA 3-ketoacyl-CoA thiolase | | 分子名称: | 3-ketoacyl-CoA thiolase, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Anderson, S.M, Skarina, T, Onopriyenko, O, Wawrzak, Z, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-03-18 | | 公開日 | 2009-03-31 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 |
|
|
3IGS
 
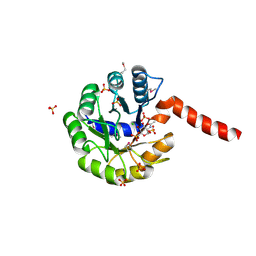 | | Structure of the Salmonella enterica N-acetylmannosamine-6-phosphate 2-epimerase | | 分子名称: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, N-acetylmannosamine-6-phosphate 2-epimerase 2, ... | | 著者 | Anderson, S.M, Wawrzak, Z, Gordon, E, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-07-28 | | 公開日 | 2009-08-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 |
|
|
4MJX
 
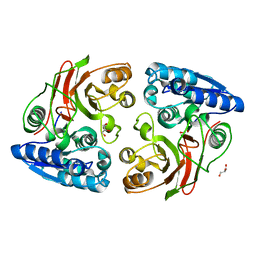 | |
4OX3
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | 分子名称: | PHOSPHATE ION, Putative carboxypeptidase YodJ, ZINC ION | | 著者 | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | 登録日 | 2014-02-04 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
3PGP
 
 | | Crystal structure of PA4794 - GNAT superfamily protein in complex with AcCoA | | 分子名称: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | 著者 | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-11-02 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
4OX5
 
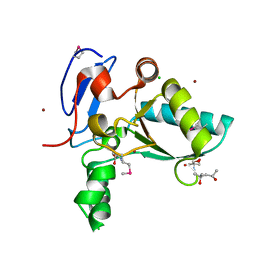 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | 登録日 | 2014-02-04 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
4OXD
 
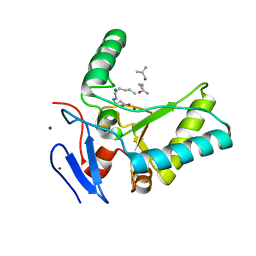 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | 分子名称: | CHLORIDE ION, LYSINE, LdcB LD-carboxypeptidase, ... | | 著者 | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | 登録日 | 2014-02-05 | | 公開日 | 2014-05-21 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
3NX3
 
 | | Crystal structure of acetylornithine aminotransferase (argD) from Campylobacter jejuni | | 分子名称: | Acetylornithine aminotransferase, MAGNESIUM ION | | 著者 | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-07-12 | | 公開日 | 2010-08-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 |
|
|
4E1L
 
 | | Crystal structure of Acetoacetyl-CoA thiolase (thlA2) from Clostridium difficile | | 分子名称: | Acetoacetyl-CoA thiolase 2, IODIDE ION | | 著者 | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-03-06 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 |
|
|
3Q58
 
 | | Structure of N-acetylmannosamine-6-Phosphate Epimerase from Salmonella enterica | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-12-27 | | 公開日 | 2011-01-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 |
|
|
191L
 
 | |
192L
 
 | |
190L
 
 | |
3K29
 
 | | Structure of a putative YscO homolog CT670 from Chlamydia trachomatis | | 分子名称: | Putative uncharacterized protein | | 著者 | Lam, R, Singer, A, Skarina, T, Onopriyenko, O, Bochkarev, A, Brunzelle, J.S, Edwards, A.M, Anderson, W.F, Chirgadze, N.Y, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-09-29 | | 公開日 | 2009-10-13 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and protein-protein interaction studies on Chlamydia trachomatis protein CT670 (YscO Homolog).
J.Bacteriol., 192, 2010
|
|
3KZL
 
 | | Crystal structure of BA2930 mutant (H183G) in complex with AcCoA | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, ... | | 著者 | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-12-08 | | 公開日 | 2009-12-22 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3N77
 
 | | Crystal structure of Idp01880, putative NTP pyrophosphohydrolase of Salmonella typhimurium LT2 | | 分子名称: | Nucleoside triphosphatase nudI | | 著者 | Frydrysiak, J, Cooper, D.R, Derewenda, U, Anderson, W.F, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-05-26 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Crystal structure of Idp01880, putative NTP pyrophosphohydrolase of Salmonella typhimurium LT2
To be Published
|
|
3N0M
 
 | | Crystal structure of BA2930 mutant (H183G) in complex with AcCoA | | 分子名称: | ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, CHLORIDE ION | | 著者 | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-05-14 | | 公開日 | 2010-06-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3N0S
 
 | | Crystal structure of BA2930 mutant (H183A) in complex with AcCoA | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, ... | | 著者 | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-05-14 | | 公開日 | 2010-06-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
