3P51
 
 | | Three-dimensional structure of protein Q2Y8N9_NITMU from nitrosospira multiformis, Northeast structural genomics consortium target NMR118 | | 分子名称: | Uncharacterized protein | | 著者 | Kuzin, A, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-10-07 | | 公開日 | 2010-10-27 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (2.056 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target NmR118
To be published
|
|
3PU2
 
 | | Crystal Structure of the Q3J4M4_RHOS4 protein from Rhodobacter sphaeroides. Northeast Structural Genomics Consortium Target RhR263. | | 分子名称: | uncharacterized protein | | 著者 | Vorobiev, S, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-12-03 | | 公開日 | 2010-12-15 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (2.606 Å) | | 主引用文献 | Crystal Structure of the Q3J4M4_RHOS4 protein from Rhodobacter sphaeroides.
To be Published
|
|
6L0Z
 
 | | The crystal structure of Salmonella enterica sugar-binding protein MalE | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-anhydro-D-glucitol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | 登録日 | 2019-09-27 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structure of Salmonella enterica sugar-binding protein MalE
To Be Published
|
|
5FZN
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | KELCH-LIKE ECH-ASSOCIATED PROTEIN 1, SULFATE ION, benzenesulfonamide | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2016-03-15 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
6L19
 
 | | The crystal structure of competence or damage-inducible protein from Enterobacter asburiae | | 分子名称: | CHLORIDE ION, GLYCEROL, PncC family amidohydrolase, ... | | 著者 | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | 登録日 | 2019-09-28 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | The crystal structure of Competence or damage-inducible protein from Enterobacter asburiae
To Be Published
|
|
6L1K
 
 | | Crystal structure of NADH-dependent butanol dehydrogenase A from Fusobacterium nucleatum | | 分子名称: | NADH-dependent butanol dehydrogenase A, PHOSPHATE ION | | 著者 | Lan, J, Shang, F, Liu, W, Xu, Y, Chen, Y. | | 登録日 | 2019-09-29 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structure of NADH-dependent butanol dehydrogenase A from Fusobacterium nucleatum
To Be Published
|
|
6L6Q
 
 | |
6LC1
 
 | |
2XR0
 
 | | Room temperature X-ray structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | 分子名称: | SULFATE ION, TOHO-1 BETA-LACTAMASE | | 著者 | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | 登録日 | 2010-09-08 | | 公開日 | 2010-12-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
1S2J
 
 | | Crystal structure of the Drosophila pattern-recognition receptor PGRP-SA | | 分子名称: | PHOSPHATE ION, Peptidoglycan recognition protein SA CG11709-PA | | 著者 | Chang, C.-I, Pili-Floury, S, Chelliah, Y, Lemaitre, B, Mengin-Lecreulx, D, Deisenhofer, J. | | 登録日 | 2004-01-08 | | 公開日 | 2004-09-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l,d-carboxypeptidase activity.
PLOS BIOL., 2, 2004
|
|
3PIN
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in complex with Trx2 | | 分子名称: | Peptide methionine sulfoxide reductase, Thioredoxin-2 | | 著者 | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | 登録日 | 2010-11-07 | | 公開日 | 2011-02-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3QV0
 
 | | Crystal structure of Saccharomyces cerevisiae Mam33 | | 分子名称: | Mitochondrial acidic protein MAM33 | | 著者 | Jiang, Y.L, Pu, Y.G, Ma, X.X, Chen, Y, Zhou, C.Z. | | 登録日 | 2011-02-24 | | 公開日 | 2011-06-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures and putative interface of Saccharomyces cerevisiae mitochondrial matrix proteins Mmf1 and Mam33.
J.Struct.Biol., 175, 2011
|
|
6M22
 
 | | KCC3 bound with DIOA | | 分子名称: | 2-[[(2~{R})-2-butyl-6,7-bis(chloranyl)-2-cyclopentyl-1-oxidanylidene-3~{H}-inden-5-yl]oxy]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | 登録日 | 2020-02-26 | | 公開日 | 2020-11-04 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
2H3R
 
 | | Crystal structure of ORF52 from Murid herpesvirus 4 (MuHV-4) (Murine gammaherpesvirus 68). Northeast Structural Genomics Consortium target MhR28B. | | 分子名称: | Hypothetical protein BQLF2 | | 著者 | Benach, J, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Cunningham, K, Ma, L.-C, Ho, C.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-05-23 | | 公開日 | 2006-08-15 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of ORF52 from Murid herpesvirus 4 (MuHV-4) (Murine gammaherpesvirus 68). Northeast Structural Genomics Consortium target MhR28B.
To be Published
|
|
5FNS
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | (3s)-{4-Chloro-3-[(N-methylmethanesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl) propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2015-11-16 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
7D4U
 
 | | ATP complex with double mutant cyclic trinucleotide synthase CdnD | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase | | 著者 | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | 登録日 | 2020-09-24 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4S
 
 | | apo-form cyclic trinucleotide synthase CdnD | | 分子名称: | Cyclic AMP-AMP-GMP synthase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION | | 著者 | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | 登録日 | 2020-09-24 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4O
 
 | | cyclic trinucleotide synthase CdnD in complex with ATP and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase, ... | | 著者 | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | 登録日 | 2020-09-24 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4J
 
 | | ddATP complex of cyclic trinucleotide synthase CdnD | | 分子名称: | 2',3'-dideoxyadenosine triphosphate, Cyclic AMP-AMP-GMP synthase, MAGNESIUM ION | | 著者 | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | 登録日 | 2020-09-24 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D48
 
 | | apo-form cyclic trinucleotide synthase CdnD | | 分子名称: | Cyclic AMP-AMP-GMP synthase, SODIUM ION | | 著者 | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | 登録日 | 2020-09-23 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
5FNQ
 
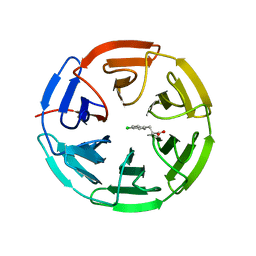 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | 3-(4-CHLOROPHENYL)PROPANOIC ACID, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2015-11-16 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
6M1Y
 
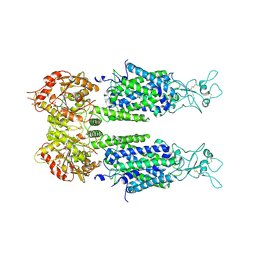 | | The overall structure of KCC3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | 登録日 | 2020-02-26 | | 公開日 | 2020-11-04 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
6M23
 
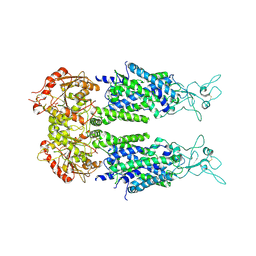 | | Overall structure of KCC2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | 登録日 | 2020-02-26 | | 公開日 | 2020-11-04 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
3Q6A
 
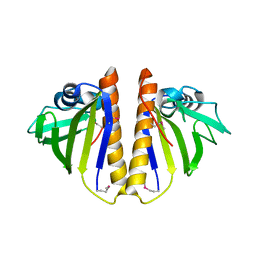 | | X-ray crystal structure of the protein SSP2350 from Staphylococcus saprophyticus, Northeast structural genomics consortium target SyR116 | | 分子名称: | uncharacterized protein | | 著者 | Seetharaman, J, Chen, Y, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-12-31 | | 公開日 | 2011-04-06 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray crystal structure of the protein SSP2350 from Staphylococcus saprophyticus, Northeast structural genomics consortium target SyR116
To be Published
|
|
7DV5
 
 | |
