3HSA
 
 | |
3H0N
 
 | |
3GO5
 
 | |
3H41
 
 | |
3IRB
 
 | |
3F1Z
 
 | |
3FMS
 
 | | Crystal structure of TM0439, a GntR transcriptional regulator | | 分子名称: | ACETATE ION, NICKEL (II) ION, Transcriptional regulator, ... | | 著者 | Zheng, M, Cooper, D.R, Yu, M, Hung, L.-W, Derewenda, U, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | 登録日 | 2008-12-22 | | 公開日 | 2009-02-10 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of Thermotoga maritima TM0439: implications for the mechanism of bacterial GntR transcription regulators with Zn2+-binding FCD domains.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3H50
 
 | |
3HBZ
 
 | |
4J1U
 
 | | Crystal structure of antibody 93F3 unstable variant | | 分子名称: | antibody 93F3 Heavy chain, antibody 93F3 Light chain | | 著者 | Wang, F. | | 登録日 | 2013-02-02 | | 公開日 | 2013-03-13 | | 最終更新日 | 2013-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Somatic hypermutation maintains antibody thermodynamic stability during affinity maturation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7KSG
 
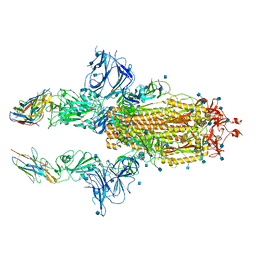 | | SARS-CoV-2 spike in complex with nanobodies E | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 glycoprotein, Spike glycoprotein | | 著者 | Hallberg, B.M, Das, H. | | 登録日 | 2020-11-22 | | 公開日 | 2021-01-20 | | 最終更新日 | 2021-06-30 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
4JRF
 
 | |
4JG5
 
 | |
4K4K
 
 | |
4MC5
 
 | | Crystal structure of a subtype H18 hemagglutinin homologue from A/flat-faced bat/Peru/033/2010 (H18N11) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | 著者 | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Stevens, J. | | 登録日 | 2013-08-21 | | 公開日 | 2013-10-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.238 Å) | | 主引用文献 | New world bats harbor diverse influenza a viruses.
Plos Pathog., 9, 2013
|
|
4MC7
 
 | | Crystal structure of a subtype N11 neuraminidase-like protein of A/flat-faced bat/Peru/033/2010 (H18N11) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE | | 著者 | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Stevens, J. | | 登録日 | 2013-08-21 | | 公開日 | 2013-10-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | New world bats harbor diverse influenza a viruses.
Plos Pathog., 9, 2013
|
|
4OCW
 
 | | Crystal structure of human Fab CAP256-VRC26.06, a potent V1V2-directed HIV-1 neutralizing antibody | | 分子名称: | CAP256-VRC26.06 heavy chain, CAP256-VRC26.06 light chain | | 著者 | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | 登録日 | 2014-01-09 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4ODH
 
 | | Crystal structure of human Fab CAP256-VRC26.UCA, a potent V1V2-directed HIV-1 neutralizing antibody | | 分子名称: | CAP256-VRC26.UCA heavy chain, CAP256-VRC26.UCA light chain | | 著者 | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | 登録日 | 2014-01-10 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.894 Å) | | 主引用文献 | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4ORG
 
 | | Crystal structure of human Fab CAP256-VRC26.04, a potent V1V2-directed HIV-1 neutralizing antibody | | 分子名称: | CAP256-VRC26.04 heavy chain, CAP256-VRC26.04 light chain | | 著者 | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | 登録日 | 2014-02-11 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.121 Å) | | 主引用文献 | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4OD1
 
 | | Crystal structure of human Fab CAP256-VRC26.03, a potent V1V2-directed HIV-1 neutralizing antibody | | 分子名称: | CAP256-VRC26.03 heavy chain, CAP256-VRC26.03 light chain | | 著者 | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | 登録日 | 2014-01-09 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4OCR
 
 | | Crystal structure of human Fab CAP256-VRC26.01, a potent V1V2-directed HIV-1 neutralizing antibody | | 分子名称: | CAP256-VRC26.01 heavy chain, CAP256-VRC26.01 light chain | | 著者 | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | 登録日 | 2014-01-09 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.895 Å) | | 主引用文献 | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
2ETS
 
 | |
2F46
 
 | |
4OD3
 
 | | Crystal structure of human Fab CAP256-VRC26.07, a potent V1V2-directed HIV-1 neutralizing antibody | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CAP256-VRC26.07 heavy chain, CAP256-VRC26.07 light chain, ... | | 著者 | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | 登録日 | 2014-01-09 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.616 Å) | | 主引用文献 | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
2HBW
 
 | |
