4P79
 
 | | Crystal structure of mouse claudin-15 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | 著者 | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | 登録日 | 2014-03-26 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
5ZFU
 
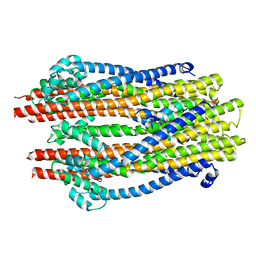 | | Structure of the ExbB/ExbD hexameric complex (ExbB6ExbD3TM) | | 分子名称: | 22-mer peptide from Biopolymer transport protein ExbD, Biopolymer transport protein ExbB | | 著者 | Yonekura, K, Yamashita, Y, Matsuoka, R, Maki-Yonekura, S. | | 登録日 | 2018-03-07 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Hexameric and pentameric complexes of the ExbBD energizer in the Ton system.
Elife, 7, 2018
|
|
5AUO
 
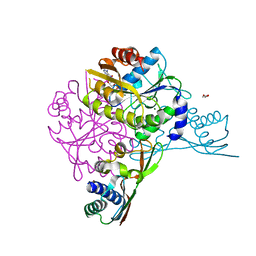 | | Crystal structure of the HypAB-Ni complex (AMPPCP) | | 分子名称: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | 著者 | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | 登録日 | 2015-05-27 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUQ
 
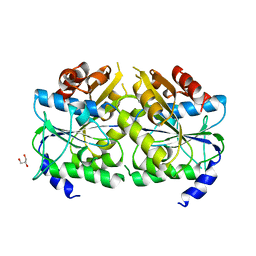 | | Crystal structure of ATPase-type HypB in the nucleotide free state | | 分子名称: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | 著者 | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | 登録日 | 2015-05-27 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.525 Å) | | 主引用文献 | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WKT
 
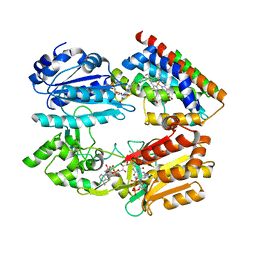 | | Complex structure of an open form of NADPH-cytochrome P450 reductase and heme oxygenase-1 | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1, ... | | 著者 | Sugishima, M, Sato, H, Higashimoto, Y, Harada, J, Wada, K, Fukuyama, K, Noguchi, M. | | 登録日 | 2013-10-31 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | Structural basis for the electron transfer from an open form of NADPH-cytochrome P450 oxidoreductase to heme oxygenase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1MSE
 
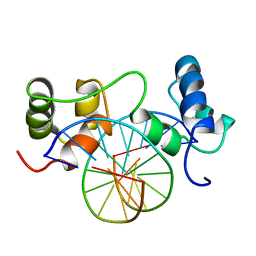 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | 分子名称: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | 著者 | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | 登録日 | 1995-01-24 | | 公開日 | 1995-03-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
1ISE
 
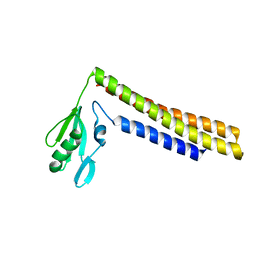 | | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly | | 分子名称: | Ribosome Recycling Factor | | 著者 | Nakano, H, Yoshida, T, Oka, S, Uchiyama, S, Nishina, K, Ohkubo, T, Kato, H, Yamagata, Y, Kobayashi, Y. | | 登録日 | 2001-11-30 | | 公開日 | 2003-10-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly
To be Published
|
|
1EWT
 
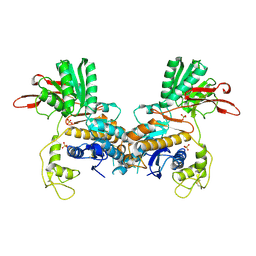 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM I | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1, SULFATE ION | | 著者 | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | 登録日 | 2000-04-27 | | 公開日 | 2000-12-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
1IW4
 
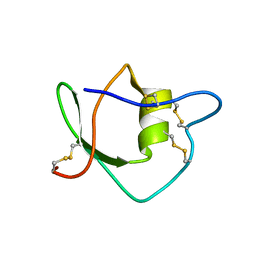 | | Solution structure of ascidian trypsin inhibitor | | 分子名称: | trypsin inhibitor | | 著者 | Hemmi, H, Yoshida, T, Kumazaki, T, Nemoto, N, Hasegawa, J, Nishioka, F, Kyogoku, Y, Yokosawa, H, Kobayashi, Y. | | 登録日 | 2002-04-19 | | 公開日 | 2002-08-28 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of ascidian trypsin inhibitor determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 41, 2002
|
|
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-15 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-15 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | 著者 | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-08 | | 公開日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
6C9H
 
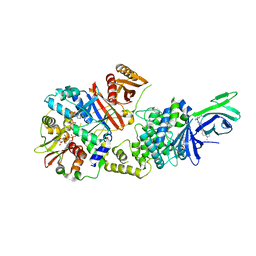 | | non-phosphorylated AMP-activated protein kinase bound to pharmacological activator R734 | | 分子名称: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | 著者 | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | 登録日 | 2018-01-26 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6C9J
 
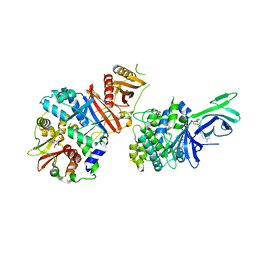 | | AMP-activated protein kinase bound to pharmacological activator R734 | | 分子名称: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | 著者 | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Brunzelle, J.S, Hitoshi, Y, Griffin, P.R, Xu, H.E, Melcher, K. | | 登録日 | 2018-01-26 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6C9G
 
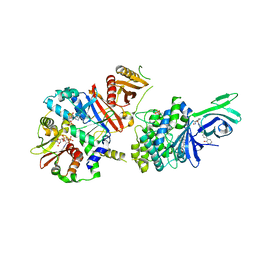 | | AMP-activated protein kinase bound to pharmacological activator R739 | | 分子名称: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | 著者 | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | 登録日 | 2018-01-26 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6C9F
 
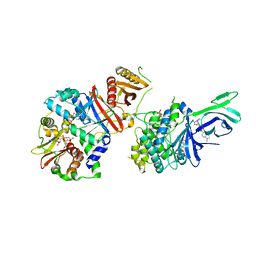 | | AMP-activated protein kinase bound to pharmacological activator R734 | | 分子名称: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | 著者 | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | 登録日 | 2018-01-26 | | 公開日 | 2018-11-28 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.924 Å) | | 主引用文献 | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
1EWV
 
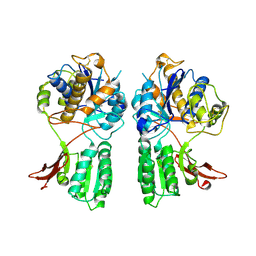 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM II | | 分子名称: | METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 | | 著者 | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | 登録日 | 2000-04-27 | | 公開日 | 2000-12-18 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
1MSF
 
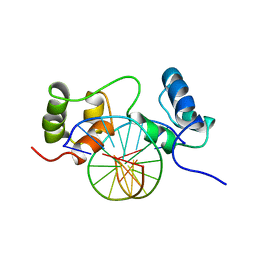 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | 分子名称: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | 著者 | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | 登録日 | 1995-01-24 | | 公開日 | 1995-03-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
8X6R
 
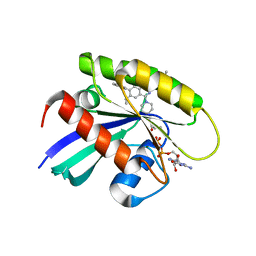 | | KRasG12C in complex with inhibitor | | 分子名称: | 1-[7-[6-ethenyl-8-ethoxy-7-(5-methyl-1~{H}-indazol-4-yl)-2-(1-methylpiperidin-4-yl)oxy-quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | 著者 | Amano, Y, Tateishi, Y. | | 登録日 | 2023-11-21 | | 公開日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Discovery of ASP6918, a KRAS G12C inhibitor: Synthesis and structure-activity relationships of 1-{2,7-diazaspiro[3.5]non-2-yl}prop-2-en-1-one derivatives as covalent inhibitors with good potency and oral activity for the treatment of solid tumors.
Bioorg.Med.Chem., 98, 2023
|
|
5DFZ
 
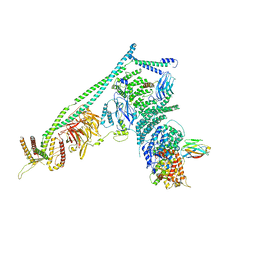 | | Structure of Vps34 complex II from S. cerevisiae. | | 分子名称: | Nanobody binding S. cerevisiae Vps34, Phosphatidylinositol 3-kinase VPS34, Putative N-terminal domain of S. cerevisiae Vps30, ... | | 著者 | Rostislavleva, K, Soler, N, Ohashi, Y, Zhang, L, Williams, R.L. | | 登録日 | 2015-08-27 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (4.4 Å) | | 主引用文献 | Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes.
Science, 350, 2015
|
|
1IY6
 
 | | Solution structure of OMSVP3 variant, P14C/N39C | | 分子名称: | OMSVP3 | | 著者 | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | 登録日 | 2002-07-23 | | 公開日 | 2003-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
1IY5
 
 | | Solution structure of wild type OMSVP3 | | 分子名称: | OMSVP3 | | 著者 | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | 登録日 | 2002-07-23 | | 公開日 | 2003-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
2SXL
 
 | | SEX-LETHAL RBD1, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | SEX-LETHAL PROTEIN | | 著者 | Inoue, M, Muto, Y, Sakamoto, H, Kigawa, T, Takio, K, Shimura, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1997-07-16 | | 公開日 | 1998-07-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal.
J.Mol.Biol., 272, 1997
|
|
5CZD
 
 | | The complex structure of VinK with VinL | | 分子名称: | 1,1'-ethane-1,2-diyldipyrrolidine-2,5-dione, 4'-PHOSPHOPANTETHEINE, Acyl-carrier-protein, ... | | 著者 | Miyanaga, A, Iwasawa, S, Shinohara, Y, Kudo, F, Eguchi, T. | | 登録日 | 2015-07-31 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structure-based analysis of the molecular interactions between acyltransferase and acyl carrier protein in vicenistatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1OED
 
 | | STRUCTURE OF ACETYLCHOLINE RECEPTOR PORE FROM ELECTRON IMAGES | | 分子名称: | Acetylcholine receptor beta subunit, Acetylcholine receptor delta subunit, Acetylcholine receptor gamma subunit, ... | | 著者 | Miyazawa, A, Fujiyoshi, Y, Unwin, N. | | 登録日 | 2003-03-24 | | 公開日 | 2003-06-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure and Gating Mechanism of the Acetylcholine Receptor Pore.
Nature, 423, 2003
|
|
